Illumina Methylation Assay
Similar to bisulfite sequencing and pyrosequencing, this method quantifies methylation levels at various loci within the genome.
Probes on the 27k array target regions of the human genome to measure methylation levels at 27,578 CpG dinucleotides in 14,495 genes.
[2] In 2016, the Infinium MethylationEPIC BeadChip ("EPIC") was released, which interrogates over 850,000 methylation sites across the human genome.
[4] For example, hypermethylation at the promoter CpG islands of a tumour suppressor gene, which in turn leads to its silencing, is frequently associated with tumourgenesis.
[4] A large scale measurement of DNA methylation patterns from a wide selection of genes may enable us to understand better the relationships between epigenetic changes and the genesis of different diseases and a better understanding of the role that epigenetics plays in tissue specific differentiation.
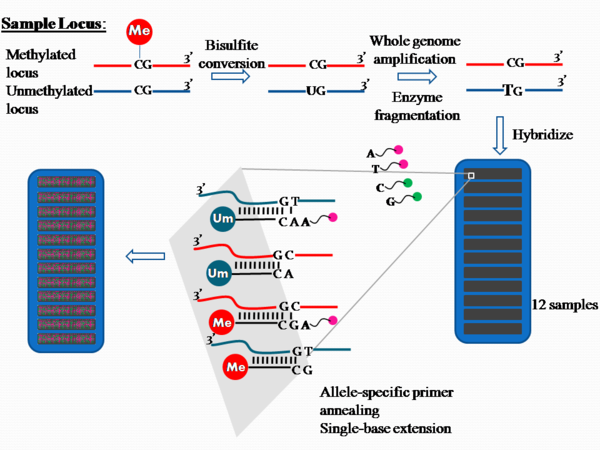
[6] Hybridization and Single-base extension On the chip, there are two bead types for each CpG (or "CG", as per Figure 1) site per locus.
[1] The raw data are analyzed by the proprietary software, and the fluorescence intensity ratios between the two bead types are calculated.