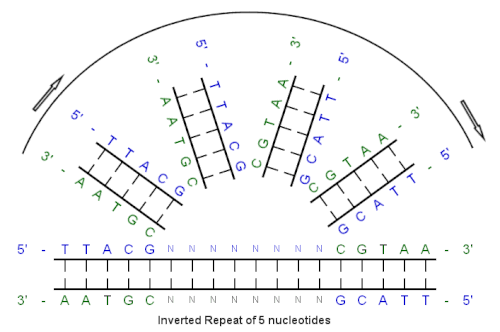
Inverted repeat
These properties play an important role in genome instability[7] and contribute not only to cellular evolution and genetic diversity[8] but also to mutation and disease.
[9] In order to study these effects in detail, a number of programs and databases have been developed to assist in discovery and annotation of inverted repeats in various genomes.
[1] The diverse genome-wide repeats are derived from transposable elements, which are now understood to "jump" about different genomic locations, without transferring their original copies.
[16] For quantification and comparison of inverted repeats between several species, namely on archaea, see [17] Pseudoknots are common structural motifs found in RNA.
There are multiple folding topologies among pseudoknots and great variation in loop lengths, making them a structurally diverse group.
[19] The ribozyme for the hepatitis delta virus (HDV) folds into a double-pseudoknot structure and self-cleaves its circular genome to produce a single-genome-length RNA.
Pseudoknots also play a role in programmed ribosomal frameshifting found in some viruses and required in the replication of retroviruses.
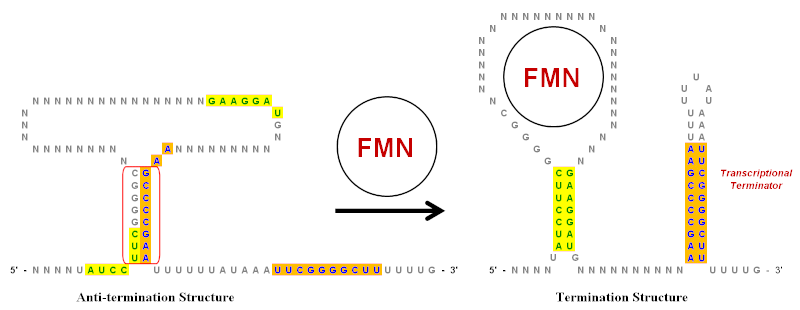
[18] Inverted repeats play an important role in riboswitches, which are RNA regulatory elements that control the expression of genes that produce the mRNA, of which they are part.
[21] This is exemplified in E. coli, where genomic sequences with long inverted repeats are seldom replicated, but rather deleted with rapidity.
[21] Again, the long inverted repeats observed in yeast greatly favor recombination within the same and adjacent chromosomes, resulting in an equally very high rate of deletion.
[21] Finally, a very high rate of deletion and recombination were also observed in mammalian chromosomes regions with inverted repeats.
[7] Thus, inverted repeats lead to special configurations in both RNA and DNA that can ultimately cause mutations and disease.
DNA in the region of the inverted repeat unwinds and then recombines, forming a four-way junction with two stem-loop structures.
During transcription, slippage and partial dissociation of the polymerase from the template strand can lead to both deletion and insertion mutations.
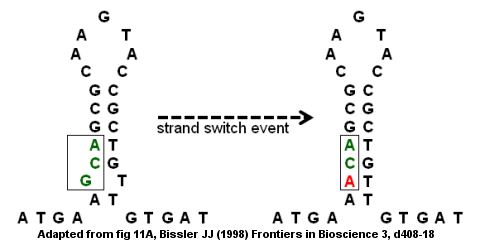
A strand switch event could result in the G (in the bump) being replaced by an A which removes the "imperfection" in the inverted repeat and provides a stronger stem-loop structure.
If the strand switch event is followed by a second round of DNA replication, the mutation may become fixed in the genome and lead to disease.
Specifically, the missense mutation would lead to a defective gene and a deficiency in antithrombin which could result in the development of venous thromboembolism (blood clots within a vein).
[9] In the illustration, a stem-loop formed from an imperfect inverted repeat is mutated with a thymine (T) nucleotide insertion as a result of an inter- or intrastrand switch.
While this addition makes the stem stronger and perfects the inverted repeat, it also creates a frameshift mutation in the nucleotide sequence which alters the reading frame and will result in an incorrect expression of the gene.