Matrix-assisted laser desorption/ionization
[1] It has been applied to the analysis of biomolecules (biopolymers such as DNA, proteins, peptides and carbohydrates) and various organic molecules (such as polymers, dendrimers and other macromolecules), which tend to be fragile and fragment when ionized by more conventional ionization methods.
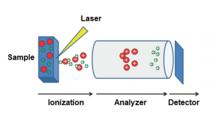
Finally, the analyte molecules are ionized by being protonated or deprotonated in the hot plume of ablated gases, and then they can be accelerated into whichever mass spectrometer is used to analyse them.
[2] The term matrix-assisted laser desorption ionization (MALDI) was coined in 1985 by Franz Hillenkamp, Michael Karas and their colleagues.
[4] The breakthrough for large molecule laser desorption ionization came in 1987 when Koichi Tanaka of Shimadzu Corporation and his co-workers used what they called the "ultra fine metal plus liquid matrix method" that combined 30 nm cobalt particles in glycerol with a 337 nm nitrogen laser for ionization.
Tanaka received one-quarter of the 2002 Nobel Prize in Chemistry for demonstrating that, with the proper combination of laser wavelength and matrix, a protein can be ionized.
[6] Karas and Hillenkamp were subsequently able to ionize the 67 kDa protein albumin using a nicotinic acid matrix and a 266 nm laser.
[8] The availability of small and relatively inexpensive nitrogen lasers operating at 337 nm wavelength and the first commercial instruments introduced in the early 1990s brought MALDI to an increasing number of researchers.
[15] A solution of one of these molecules is made, often in a mixture of highly purified water and an organic solvent such as acetonitrile (ACN) or ethanol.
The identification of suitable matrix compounds is determined to some extent by trial and error, but they are based on some specific molecular design considerations.
[16] They have a strong optical absorption in either the UV or IR range,[17] so that they rapidly and efficiently absorb the laser irradiation.
Co-crystallization is a key issue in selecting a proper matrix to obtain a good quality mass spectrum of the analyte of interest.
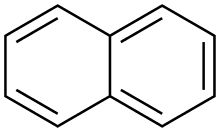
As mentioned above, acid-base like reactions are often utilized to ionize the sample, however, molecules with conjugated pi systems, such as naphthalene like compounds, can also serve as an electron acceptor and thus a matrix for MALDI/TOF.
[22] There are several variations of the MALDI technology and comparable instruments are today produced for very different purposes, from more academic and analytical, to more industrial and high throughput.
[27] IR-MALDI also has the advantage of greater material removal (useful for biological samples), less low-mass interference, and compatibility with other matrix-free laser desorption mass spectrometry methods.
The TOF measurement procedure is also ideally suited to the MALDI ionization process since the pulsed laser takes individual 'shots' rather than working in continuous operation.
In contrast, ionization at atmopsheric pressure can generate highly-charged analytes as was first shown for infrared [35] and later also for nitrogen lasers.
[36] Multiple charging of analytes is of great importance, because it allows to measure high-molecular-weight compounds like proteins in instruments, which provide only smaller m/z detection ranges such as quadrupoles.
The lucky survivor model (cluster ionization mechanism[2]) postulates that analyte molecules are incorporated in the matrix maintaining the charge state from solution.
[47] The matrix-assisted ionization (MAI) method uses matrix preparation similar to MALDI but does not require laser ablation to produce analyte ions of volatile or nonvolatile compounds.
[48] Simply exposing the matrix with analyte to the vacuum of the mass spectrometer creates ions with nearly identical charge states to electrospray ionization.
[51] The issue of low ion yields had been addressed, already shortly after introduction of MALDI by various attempts, including post-ionization utilizing a second laser.
This might be attributed to the fact that axial time-of-flight instruments were used, which operate at pressures in the source region of 10−5 to 10−6, which results in rapid plume expansion with particle velocities of up to 1000 m/s.
[53] In 2015, successful laser post-ionization was reported, using a modified MALDI source operated at an elevated pressure of ~3 mbar coupled to an orthogonal time-of-flight mass analyzer, and employing a wavelength-tunable post-ionization laser, operated at wavelength from 260 nm to 280 nm, below the two-photon ionization threshold of the matrices used, which elevated ion yields of several lipids and small molecules by up to three orders of magnitude.
[54] This approach, called MALDI-2, due to the second laser, and the second MALDI-like ionization process, was afterwards adopted for other mass spectrometers, all equipped with sources operating in the low mbar range.
[61] THAP,[62] DHAP,[63] and a mixture of 2-aza-2-thiothymine and phenylhydrazine[64] have been identified as matrices that could be used to minimize loss of sialic acid during MALDI MS analysis of glycosylated peptides.
[70] Polymers with polydispersity greater than 1.2 are difficult to characterize with MALDI due to the signal intensity discrimination against higher mass oligomers.
The mass spectra of expressed proteins generated are analyzed by dedicated software and compared with stored profiles for species determination in what is known as biotyping.
It offers benefits to other immunological or biochemical procedures and has become a common method for species identification in clinical microbiological laboratories.
[78] One main advantage over other microbiological identification methods is its ability to rapidly and reliably identify, at low cost, a wide variety of microorganisms directly from the selective medium used to isolate them.
[92] Impaired cellular signaling due to mutations in membrane proteins has been long suspected to contribute to pancreatic cancer.