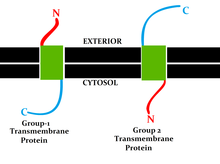
Membrane topology
[12][13][14][15] Unsupervised learning methods are based on the principle that topology depends on the maximum divergence of the amino acid distributions in different structural parts.
[16][17] It was also shown that locking a segment location based on prior knowledge about the structure improves the prediction accuracy.
[20] HTP database[21][22] provides a collection of topologies that are computationally predicted for human transmembrane proteins.
Discrimination of signal peptides and transmembrane segments is an additional problem in topology prediction treated with a limited success by different methods.
By predicting signal peptides and transmembrane helices simultaneously (Phobius[14]), the errors caused by cross-prediction are reduced and the performance is substantially increased.