Modularity (networks)
Modularity is often used in optimization methods for detecting community structure in networks.
Biological networks, including animal brains, exhibit a high degree of modularity.
However, modularity maximization is not statistically consistent, and finds communities in its own null model, i.e. fully random graphs, and therefore it cannot be used to find statistically significant community structures in empirical networks.
Furthermore, it has been shown that modularity suffers a resolution limit and, therefore, it is unable to detect small communities.
Many scientifically important problems can be represented and empirically studied using networks.
Modularity is one such measure, which when maximized, leads to the appearance of communities in a given network.
The value of the modularity for unweighted and undirected graphs lies in the range
[1] In the most common version of the concept, the randomization of the edges is done so as to preserve the degree of each vertex.
links (edges) such that the graph can be partitioned into two communities using a membership variable
(It is important to note that multiple edges may exist between two nodes, but here we assess the simplest case).
The expected number of edges shall be computed using the concept of a configuration model.
Thus, even though the node degree distribution of the graph remains intact, the configuration model results in a completely random network.
We calculate the expected number of full edges between these nodes.
, so the expected value of this quantity is Many texts then make the following approximations, for random networks with a large number of edges.
Additionally, in a large random network, the number of self-loops and multi-edges is vanishingly small.
[5] Ignoring self-loops and multi-edges allows one to assume that there is at most one edge between any two nodes.
becomes a binary indicator variable, so its expected value is also the probability that it equals
, which means one can approximate the probability of an edge existing between nodes
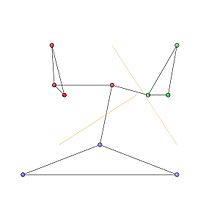
The communities in the graph are represented by the red, green and blue node clusters in Fig 1.
[1] This function has the same form as the Hamiltonian of an Ising spin glass, a connection that has been exploited to create simple computer algorithms, for instance using simulated annealing, to maximize the modularity.
The general form of the modularity for arbitrary numbers of communities is equivalent to a Potts spin glass and similar algorithms can be developed for this case also.
Because of this, the method cannot be used to reliably obtain statistically significant community structure in empirical networks.
Moreover, this implies that the expected number of edges between two groups of nodes decreases if the size of the network increases.
So, if a network is large enough, the expected number of edges between two groups of nodes in modularity's null model may be smaller than one.
So, even weakly interconnected complete graphs, which have the highest possible density of internal edges, and represent the best identifiable communities, would be merged by modularity optimization if the network were sufficiently large.
[10] For this reason, optimizing modularity in large networks would fail to resolve small communities, even when they are well defined.
This bias is inevitable for methods like modularity optimization, which rely on a global null model.
[11] There are two main approaches which try to solve the resolution limit within the modularity context: the addition of a resistance r to every node, in the form of a self-loop, which increases (r>0) or decreases (r<0) the aversion of nodes to form communities;[12] or the addition of a parameter γ>0 in front of the null-case term in the definition of modularity, which controls the relative importance between internal links of the communities and the null model.
[7] Optimizing modularity for values of these parameters in their respective appropriate ranges, it is possible to recover the whole mesoscale of the network, from the macroscale in which all nodes belong to the same community, to the microscale in which every node forms its own community, hence the name multiresolution methods.
[13] There are a couple of software tools available that are able to compute clusterings in graphs with good modularity.