Molecular mechanics
The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using force fields.
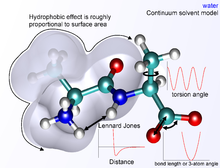
The dihedral or torsional terms typically have multiple minima and thus cannot be modeled as harmonic oscillators, though their specific functional form varies with the implementation.
Description of van der Waals forces by the Lennard-Jones 6–12 potential introduces inaccuracies, which become significant at short distances.
A variety of methods are used to address this problem, the simplest being a cutoff radius similar to that used for the van der Waals terms.
Other more sophisticated but computationally intensive methods are particle mesh Ewald (PME) and the multipole algorithm.
Given enough sampling and subject to the ergodic hypothesis, molecular dynamics trajectories can be used to estimate thermodynamic parameters of a system or probe kinetic properties, such as reaction rates and mechanisms.
This method uses an appropriate algorithm (e.g. steepest descent) to find the molecular structure of a local energy minimum.
At finite temperature, the molecule spends most of its time in these low-lying states, which thus dominate the molecular properties.
A variety of water models exist with increasing levels of complexity, representing water as a simple hard sphere (a united-atom model), as three separate particles with fixed bond angle, or even as four or five separate interaction centers to account for unpaired electrons on the oxygen atom.
This method is useful to prevent artifacts that arise from vacuum simulations and reproduces bulk solvent properties well, but cannot reproduce situations in which individual water molecules create specific interactions with a solute that are not well captured by the solvent model, such as water molecules that are part of the hydrogen bond network within a protein.