Force field (chemistry)
More precisely, the force field refers to the functional form and parameter sets used to calculate the potential energy of a system on the atomistic level.
The parameters for a chosen energy function may be derived from classical laboratory experiment data, calculations in quantum mechanics, or both.
[1] A large number of different force field types exist today (e.g. for organic molecules, ions, polymers, minerals, and metals).
For a component-specific parametrization, the considered force field is developed solely for describing a single given substance (e.g.
[3] A different important differentiation addresses the physical structure of the models: All-atom force fields provide parameters for every type of atom in a system, including hydrogen, while united-atom interatomic potentials treat the hydrogen and carbon atoms in methyl groups and methylene bridges as one interaction center.
[4][5] Coarse-grained potentials, which are often used in long-time simulations of macromolecules such as proteins, nucleic acids, and multi-component complexes, sacrifice chemical details for higher computing efficiency.
Additionally, "improper torsional" terms may be added to enforce the planarity of aromatic rings and other conjugated systems, and "cross-terms" that describe the coupling of different internal variables, such as angles and bond lengths.
A large number of force fields based on this or similar energy expressions have been proposed in the past decades for modeling different types of materials such as molecular substances, metals, glasses etc.
As it is rare for bonds to deviate significantly from their equilibrium values, the most simplistic approaches utilize a Hooke's law formula:
Though the formula of Hooke's law provides a reasonable level of accuracy at bond lengths near the equilibrium distance, it is less accurate as one moves away.
[4][9] However, for most practical applications these differences are negligible, and inaccuracies in predictions of bond lengths are on the order of the thousandth of an angstrom, which is also the limit of reliability for common force fields.
Nevertheless, the term 'empirical' is often used in the context of force field parameters when macroscopic material property data was used for the fitting.
[3][22][23] The assignment of atomic charges often follows quantum mechanical protocols with some heuristics, which can lead to significant deviation in representing specific properties.
[24][25][26] A large number of workflows and parametrization procedures have been employed in the past decades using different data and optimization strategies for determining the force field parameters.
The parameters for molecular simulations of biological macromolecules such as proteins, DNA, and RNA were often derived/transferred from observations for small organic molecules, which are more accessible for experimental studies and quantum calculations.
In recent years, some databases have attempted to collect, categorize and make force fields digitally available.
For example, the openKim database focuses on interatomic functions describing the individual interactions between specific elements.
[30] The TraPPE database focuses on transferable force fields of organic molecules (developed by the Siepmann group).
[5][32] Functional forms and parameter sets have been defined by the developers of interatomic potentials and feature variable degrees of self-consistency and transferability.
[34] The use of accurate representations of chemical bonding, combined with reproducible experimental data and validation, can lead to lasting interatomic potentials of high quality with much fewer parameters and assumptions in comparison to DFT-level quantum methods.
[37] The remedy is that point charges have a clear interpretation[26] and virtual electrons can be added to capture essential features of the electronic structure, such additional polarizability in metallic systems to describe the image potential, internal multipole moments in π-conjugated systems, and lone pairs in water.
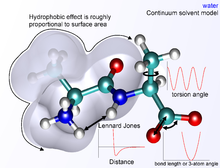
However, application of one value of dielectric constant is a coarse approximation in the highly heterogeneous environments of proteins, biological membranes, minerals, or electrolytes.
A more general theory of van der Waals forces in condensed media was developed by A. D. McLachlan in 1963 and included the original London's approach as a special case.
[45] This is in contrast to combinatorial rules or Slater-Kirkwood equation applied for development of the classical force fields.
The major underlying challenge is the huge conformation space of polymeric molecules, which grows beyond current computational feasibility when containing more than ~20 monomers.
[47] Force fields have been applied successfully for protein structure refinement in different X-ray crystallography and NMR spectroscopy applications, especially using program XPLOR.
These shortcomings are related to interatomic potentials and to the inability to sample the conformation space of large molecules effectively.
[41] The parameters of proteins force fields reproduce the enthalpy of sublimation, i.e., energy of evaporation of molecular crystals.
However, protein folding and ligand binding are thermodynamically closer to crystallization, or liquid-solid transitions as these processes represent freezing of mobile molecules in condensed media.
Indeed, the energies of H-bonds in proteins are ~ -1.5 kcal/mol when estimated from protein engineering or alpha helix to coil transition data,[64][65] but the same energies estimated from sublimation enthalpy of molecular crystals were -4 to -6 kcal/mol,[66] which is related to re-forming existing hydrogen bonds and not forming hydrogen bonds from scratch.