Phagosome
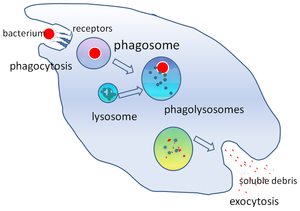
Phagosomes are formed when pathogens or opsonins bind to a transmembrane receptor, which are randomly distributed on the phagocyte cell surface.
Upon binding, "outside-in" signalling triggers actin polymerisation and pseudopodia formation, which surrounds and fuses behind the microorganism.
It is part of the adaptive immune system, but it links to the innate response by recruiting macrophages to phagocytose pathogens.
Complement-mediated internalisation has much less significant membrane protrusions, but the downstream signalling of both pathways converge to activate Rho GTPases.
Phagosomes can engulf artificial low-density latex beads and then purified along a sucrose concentration gradient, allowing the structure and composition to be studied.
The different enzymes function at various optimal pH, forming a range so they each work in narrow stages of the maturation process.
The phagosome moves along microtubules of the cytoskeleton, fusing with endosomes and lysosomes sequentially in a dynamic "kiss-and-run" manner.
[17] Vacuolar proton pumps (v-ATPase) are delivered to the phagosome to acidify the organelle compartment, creating a more hostile environment for pathogens and facilitating protein degradation.
Mycobacterium tuberculosis has a very hydrophobic cell wall, which is hypothesised to prevent membrane recycling and recruitment of fusion factors, so the phagosome does not fuse with lysosomes and the bacterium avoids degradation.
[8] Shortly after internalisation, F-actin depolymerises from the newly formed phagosome so it becomes accessible to endosomes for fusion and delivery of proteins.
[8] The maturation process is divided into early and late stages depending on characteristic protein markers, regulated by small Rab GTPases.
[19] The exact maturation pathway in mammals is not well understood, but it is suggested that HOPS can bind Rab7 and displace the guanosine nucleotide dissociation inhibitor (GDI).
The compartment is also acidic due to proton pumps (v-ATPases) that transport H+ across the membrane, used to denature the bacterial proteins.
The granules contain NADPH oxidase and myeloperoxidase, which produce toxic oxygen and chlorine derivatives to kill pathogens in an oxidative burst.
[9] The two proteins, along with Rho GTPases, are important components of the innate immune response, inducing cytokine production and activating the MAP kinase signalling cascade.
They need to retain protein fragments of a suitable size for specific bacterial recognition, so the peptides are only partially degraded.
[24] For the soil amoeba Dictyostelium discoideum, their main food source is the bacteria Legionella pneumophila, which causes Legionnaire's disease in humans.
Inflammation is only triggered by certain pathogen- or damage-associated molecular patterns (PAMPs or DAMPs), the removal of senescent cells is non-inflammatory.
[14] Autophagosomes are different from phagosomes in that they are mainly used to selectively degrade damaged cytosolic organelles such as mitochondria (mitophagy).
[27] Autophagy is not limited to professional phagocytes, it is first discovered in rat hepatocytes by cell biologist Christian de Duve.