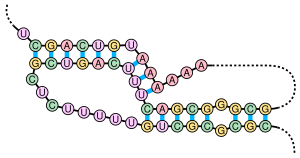
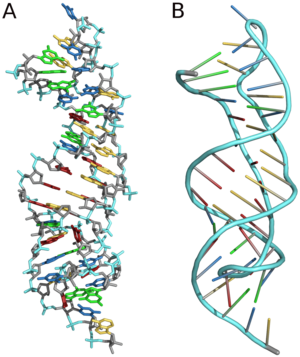
Pseudoknot
The structural configuration of pseudoknots does not lend itself well to bio-computational detection due to its context-sensitivity or "overlapping" nature.
It is possible to identify a limited class of pseudoknots using dynamic programming, but these methods are not exhaustive and scale worse as a function of sequence length than non-pseudoknotted algorithms.
[3][4] The general problem of predicting lowest free energy structures with pseudoknots has been shown to be NP-complete.
[5][6] Several important biological processes rely on RNA molecules that form pseudoknots, which are often RNAs with extensive tertiary structure.
To reflect this difference, pseudoknots are classed into H-, K-, L-, M-types, with each successive type adding a layer of step intercalation.