Quantitative genetics
[1]: 27–69 Mendel himself had to discuss this matter in his famous paper,[2] especially with respect to his peas' attribute tall/dwarf, which actually was derived by adding a cut-off point to "length of stem".
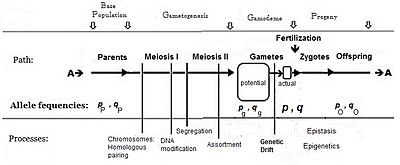
Panmixia rarely actually occurs in nature,[16]: 152–180 [17] as gamete distribution may be limited, for example by dispersal restrictions or by behaviour, or by chance sampling (those local perturbations mentioned above).
Only the latter sets the definitive frequencies for the zygotes: this is the true "gamodeme" ("gamo" refers to the gametes, and "deme" derives from Greek for "population").
Following completion of these five binomial sampling events, the resultant actual gamodemes each contained different allele frequencies—(pk and qk).
These results reveal the fact that the alleles most prevalent in the "gene-pool" (also called the "germplasm") determine performance, not the level of homozygosity per se.
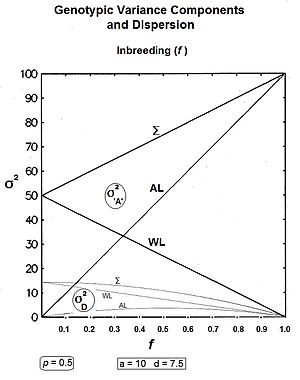
The graphs to the left show levels of inbreeding over twenty generations arising from genetic drift for various actual gamodeme sizes (2N).
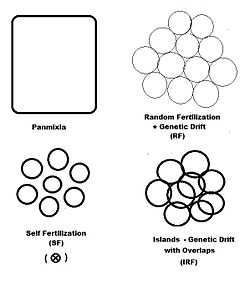
In previous sections, dispersive random fertilization (genetic drift) has been considered comprehensively, and self-fertilization and hybridizing have been examined to varying degrees.
The bulk population consists of weighted averages of sample sizes, allele and genotype frequencies and progeny means, as was done for genetic drift in an earlier section.
A parallel reasoning can be applied to the lower part of the diagram, taking care with the differences in frequencies and gene effects.
However, it is a serious non sequitur in logic to regard them as accounting for the dominance (heterozygosis) in the entire gene model : they are simply functions of "d" and not an audit of the "d" in the system.
Until this mono-genic treatment is replaced by a multi-genic one, and until epistasis is resolved in the light of the findings of epigenetics, the Genotypic variance has only the components considered here.
Most of these components are affected by the change of central focus from homozygote mid-point (mp) to population mean (G), the latter being the basis of the Correction Factor.
However, this was found subsequently to under-estimate slightly the total Genic variance, and a new variance-based derivation led to a refined version.
Even the "external" environment can be divided into spatial and temporal components ("Sites" and "Years"); or into partitions such as "litter" or "family", and "culture" or "history".
Fisher proposed that this narrow-sense heritability might be appropriate in considering the results of natural selection, focusing as it does on change-ability, that is upon "adaptation".
This value can only be determined for traits that manifest multiple times in the organism's lifetime, such as adult body mass, metabolic rate or litter size.
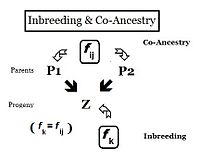
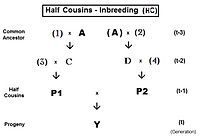
This can be viewed as the probability that two random gametes from ancestor A carry autozygous alleles, and in that context is called the coefficient of parentage ( fAA ).
Following the "B" path, the probability that any autozygous allele is "passed on" to each successive parent is again (1/2) at each step (including the last one to the "target" X ).
Recall that fAA and fBB were defined earlier (in Pedigree analysis) as coefficients of parentage, equal to (1/2)[1+fA ] and (1/2)[1+fB ] respectively, in the present context.
After substitution with corresponding inbreeding coefficients, gathering of terms and simplifying, this becomes ft = (1/4) [ 3 f(t-1) + (1/4) [2 f(t-2) + f(t-3) + 1 ]] , which is a version for iteration—useful for observing the general pattern, and for computer programming.
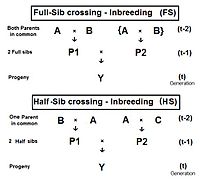
The progeny-group of a harem with a dominant male, for example, may contain elements of sib-crossing, cousin crossing, and backcrossing, as well as genetic drift, especially of the "island" type.
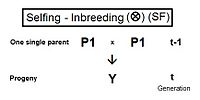
This maintenance may be through selfing, or through full-sib or half-sib crossing, or through restricted randomly fertilized populations, depending on the species' reproductive possibilities.
This formula arose because of the rules imposed by sexual reproduction: (i) two parents contributing virtually equal shares of autosomal genes, and (ii) successive dilution for each generation between the zygote and the "focus" level of parentage.
Therefore, an unadjusted sum of cross-products (USCP)—using all products { parent-pair-frequency * mid-parent-gene-effect * offspring-genotype-mean }—is adjusted by subtracting the {overall genotypic mean}2 as correction factor (CF).
[13] : 132–141 [14] : 134–147 The most obvious application is an experiment that contains all parents and their offspring, with or without reciprocal crosses, preferably replicated without bias, enabling estimation of all appropriate means, variances and covariances, together with their standard errors.
[9][49][58][59] The regression coefficient estimates the rate of change of the function predicting Y from X, based on minimizing the residuals between the fitted curve and the observed data (MINRES).
Therefore, an unadjusted sum of cross-products (USCP) using all products—{ parent-pair-frequency * the square of the offspring-genotype-mean }—is adjusted by subtracting the {overall genotypic mean}2 as correction factor (CF).
By taking partial integrations over curve-sections of interest, and some rearranging of the algebra, it can be shown that the "selection differential" is S = [ y (σ / Prob.)]
[67]: 123–124 The latter reference also gives values of i adjusted for small populations (400 and less),[67]: 111–122 where "quasi-infinity" cannot be assumed (but was presumed in the "Normal Distribution" outline above).
Referral to the section on "Genetic drift" brings to mind that it also effects changes in allele and genotype frequencies, and associated means; and that this is the companion aspect to the dispersion considered here ("the other side of the same coin").