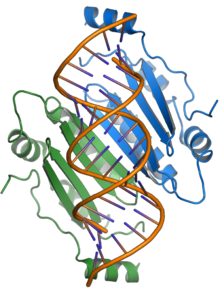
RNA silencing suppressor p19
[1][2][3] The p19 protein is considered a significant virulence factor[4] and a component of an evolutionary arms race between plants and their pathogens.
The p19 protein is also capable of binding to microRNA molecules that are endogenous to the host cell, as well as the siRNAs that are ultimately derived from the virus's own genome.
Notably, an exception to this pattern is p19's inefficiency in interacting with the microRNA miR-168, a regulatory non-coding RNA that represses expression of argonaute-1 (AGO1).
[11][12] Sequestration of dsRNA is a common viral counter-defense strategy against RNA silencing, evolved in a form of evolutionary arms race between virus and host.
The open reading frame encoding p19 was originally discovered in the late 1980s when the virus's genome was sequenced; it was subsequently demonstrated that the predicted protein was indeed expressed from the gene, although its role in promoting virulence and infectivity was initially underappreciated.