Restriction fragment length polymorphism
RFLP analysis was an important early tool in genome mapping, localization of genes for genetic disorders, determination of risk for disease, and paternity testing.
The DNA fragments produced by the digest are then separated by length through a process known as agarose gel electrophoresis and transferred to a membrane via the Southern blot procedure.
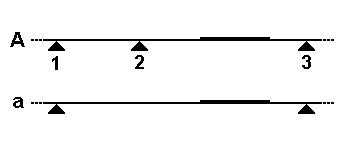
In allele A, the genome is cleaved by a restriction enzyme at three nearby sites (triangles), but only the rightmost fragment will be detected by the probe.
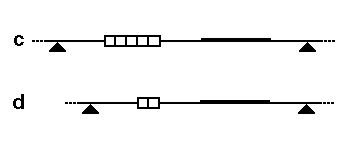
The second diagram shows how this fragment size variation would look on a Southern blot, and how each allele (two per individual) might be inherited in members of a family.
[1] The results of the Human Genome Project have largely replaced the need for RFLP mapping, and the identification of many single-nucleotide polymorphisms (SNPs) in that project (as well as the direct identification of many disease genes and mutations) has replaced the need for RFLP disease linkage analysis (see SNP genotyping).
Terminal restriction fragment length polymorphism (TRFLP or sometimes T-RFLP) is a technique initially developed for characterizing bacterial communities in mixed-species samples.
A number of different software tools have been developed to automate the process of band matching, comparison and data basing of TRFLP profiles.
Alternatively, the amplified segment can be analyzed by allele-specific oligonucleotide (ASO) probes, a process that can often be done by a simple dot blot.