SNP genotyping
Because SNPs are conserved during evolution, they have been proposed as markers for use in quantitative trait loci (QTL) analysis and in association studies in place of microsatellites.
[1] The increase of interest in SNPs has been reflected by the furious development of a diverse range of SNP genotyping methods.
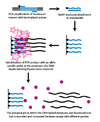
[1] Dynamic allele-specific hybridization (DASH) genotyping takes advantage of the differences in the melting temperature in DNA that results from the instability of mismatched base pairs.
[citation needed] In the first step, a genomic segment is amplified and attached to a bead through a PCR reaction with a biotinylated primer.
Other benefits of DASH include its ability to work with label free probes and its simple design and performance conditions.
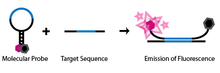
[citation needed] SNP detection through molecular beacons makes use of a specifically engineered single-stranded oligonucleotide probe.
This conformational change permits the fluorophore and quencher to be free of their tight proximity due to the hairpin association, allowing the molecule to fluoresce.
By comparing the differential amount of hybridization of the target DNA to each of these redundant probes, it is possible to determine specific homozygous and heterozygous alleles.
The Affymetrix Human SNP 5.0 GeneChip performs a genome-wide assay that can genotype over 500,000 human SNPs (Affymetrix 2007).. A broad range of enzymes including DNA ligase, DNA polymerase and nucleases have been employed to generate high-fidelity SNP genotyping methods.
By performing a digestion on a genomic sample and determining fragment lengths through a gel assay it is possible to ascertain whether or not the enzymes cut the expected restriction sites.
A failure to cut the genomic sample results in an identifiably larger than expected fragment implying that there is a mutation at the point of the restriction site which is rendering it protection from nuclease activity.
The two primer pairs are also designed such that their PCR products are of a significantly different length allowing for easily distinguishable bands by gel electrophoresis or melt temperature analysis.
An alternative strategy is to run multiple qPCR reactions with different primer sets that target each allele separately.
To achieve high enough specificity, the primer sequence may require placement of an artificial mismatch near its 3'-end, which is an approach generally known as Taq-MAMA.
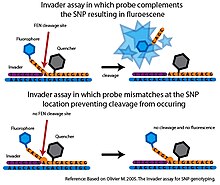
If the SNP nucleotide in the target DNA is not complementary to the allele-specific probe, the correct tripartite structure is not formed and no cleavage occurs.
The Invader assay is usually coupled with fluorescence resonance energy transfer (FRET) system to detect the cleavage event.
However, in its original format, only one SNP allele could be interrogated per reaction sample and it required a large amount of target DNA to generate a detectable signal in a reasonable time frame.
These high-throughput platforms have not progressed beyond the proof-of-principle stage and so far the Invader system has not been used in any large scale SNP genotyping projects.
These formats use a wide range of detection techniques that include MALDI-TOF Mass spectrometry (see Sequenom) and ELISA-like methods.
Broadly referred to as arrayed primer extension (APEX), this technology has several benefits over methods based on differential hybridization of probes.
[1] Illumina Incorporated's Infinium assay is an example of a whole-genome genotyping pipeline that is based on primer extension method.
The assay requires forward and reverse PCR primers that will amplify a region that includes the SNP polymorphic site.
Generally, TaqMan is limited to applications that involve interrogating a small number of SNPs since optimal probes and reaction conditions must be designed for each SNP (Syvanen 2001).
[citation needed] The characteristic DNA properties of melting temperature and single stranded conformation have been used in several applications to distinguish SNP alleles.
When applied to a gel, the tertiary shape will determine the mobility of the ssDNA, providing a mechanism to differentiate between SNP alleles.
Either you position the primers very close to either side of the SNP in question (small amplicon genotyping, Liew, 2004) or amplify a larger region (100-400bp in length) for scanning purposes.
Heterozygotes are even easier to differentiate because they have heteroduplexes generated (refer to the gel-based explanations) which broadens the melt transition and usually gives two discernible peaks.
Numerous investigators have been able to successfully eliminate the majority of their sequencing through melt-based scanning, allowing accurate locus-based genotyping of large numbers of individuals.
[9] Many investigators have found scanning for mutations using high resolution melting as a viable and practical way to study entire genes.
However, their ability to generate results in real-time and their potential to be massively scaled up makes them a viable option for sequencing small regions to perform SNP genotyping.