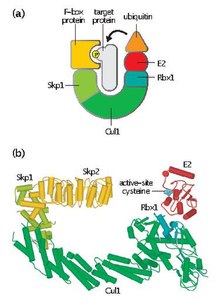
SCF complex
[1] Along with the anaphase-promoting complex,[2] SCF has important roles in the ubiquitination of proteins involved in the cell cycle.
Temperature-sensitive cell division cycle (Cdc) mutants—such as Cdc4, Cdc34, and Cdc53[6]—arrested in G1 with unreplicated DNA and multiple elongated buds.
Next, biochemical studies revealed that Cdc34 is an E2 enzyme that physically interacts with an E3 ubiquitin ligase complex containing Skp1, Cdc4, and several other proteins.
Subsequent genetic studies in Caenorhabditis elegans later contributed to the elucidation of other SCF complex components.
[8] The eukaryotic cell cycle[9] is regulated through the synthesis, degradation, binding interactions, post-translational modifications of regulatory proteins.
Fbw7, which is the human homolog of cdc4 in yeast, is an FBP that targets Cyclin E, Myc, Notch and c-Jun for degradation.
Fbw7 is known to be a haplo-insufficient tumor suppressor gene implicated in several sporadic carcinomas, for which one mutant allele is enough to disturb the wild type phenotype.
[16][17] Mutations that prevent phosphorylation of Cyclin F alter the activity of SCF-Cyclin F, which likely affects downstream processes pertinent to neuron degeneration in ALS and FTD.
Recently, SCF complexes have become an attractive anti-cancer target because of their upregulation in some human cancers and their biochemically distinct active sites.