Single-linkage clustering
[1] However, it is popular in astronomy for analyzing galaxy clusters, which may often involve long strings of matter; in this application, it is also known as the friends-of-friends algorithm.
At each step, the two clusters separated by the shortest distance are combined.
The function used to determine the distance between two clusters, known as the linkage function, is what differentiates the agglomerative clustering methods.
The shortest of these pairwise distances that remain at any step causes the two clusters whose elements are involved to be merged.
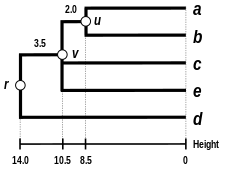
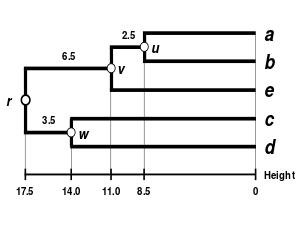
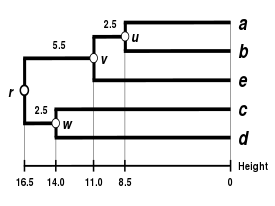
The result of the clustering can be visualized as a dendrogram, which shows the sequence in which clusters were merged and the distance at which each merge took place.
[3] Mathematically, the linkage function – the distance D(X,Y) between clusters X and Y – is described by the expression where X and Y are any two sets of elements considered as clusters, and d(x,y) denotes the distance between the two elements x and y.
The following algorithm is an agglomerative scheme that erases rows and columns in a proximity matrix as old clusters are merged into new ones.
The single linkage algorithm is composed of the following steps: This working example is based on a JC69 genetic distance matrix computed from the 5S ribosomal RNA sequence alignment of five bacteria: Bacillus subtilis (
(see the final dendrogram) We then proceed to update the initial proximity matrix
(see below), reduced in size by one row and one column because of the clustering of a with b.
are not affected by the matrix update as they correspond to distances between elements not involved in the first cluster.
We now reiterate the three previous actions, starting from the new distance matrix
Because of the ultrametricity constraint, the branches joining a or b to v, and c to v, and also e to v are equal and have the following total length: We deduce the missing branch length: We then proceed to update the
(see below), reduced in size by two rows and two columns because of the clustering of
The naive algorithm for single linkage clustering is essentially the same as Kruskal's algorithm for minimum spanning trees.
However, in single linkage clustering, the order in which clusters are formed is important, while for minimum spanning trees what matters is the set of pairs of points that form distances chosen by the algorithm.
In the naive algorithm for agglomerative clustering, implementing a different linkage scheme may be accomplished simply by using a different formula to calculate inter-cluster distances in the algorithm.
The formula that should be adjusted has been highlighted using bold text in the above algorithm description.
However, more efficient algorithms such as the one described below do not generalize to all linkage schemes in the same way.
The naive algorithm for single-linkage clustering is easy to understand but slow, with time complexity
[6] In 1973, R. Sibson proposed an algorithm with time complexity
The slink algorithm represents a clustering on a set of
These functions are both determined by finding the smallest cluster
Storing these functions in two arrays that map each item number to its function value takes space
As Sibson shows, when a new item is added to the set of items, the updated functions representing the new single-linkage clustering for the augmented set, represented in the same way, can be constructed from the old clustering in time
The SLINK algorithm then loops over the items, one by one, adding them to the representation of the clustering.
[7][8] An alternative algorithm, running in the same optimal time and space bounds, is based on the equivalence between the naive algorithm and Kruskal's algorithm for minimum spanning trees.
Instead of using Kruskal's algorithm, one can use Prim's algorithm, in a variation without binary heaps that takes time
to construct the minimum spanning tree (but not the clustering) of the given items and distances.
Then, applying Kruskal's algorithm to the sparse graph formed by the edges of the minimum spanning tree produces the clustering itself in an additional time