Single-molecule experiment
Today, systems investigated using single-molecule techniques include the movement of myosin on actin filaments in muscle tissue and the spectroscopic details of individual local environments in solids.
In addition, one of the earliest means of detecting single molecules, came about in the field of ion channels with the development of the patch clamp technique by Erwin Neher and Bert Sakmann (who later went on to win the Nobel prize for their seminal contributions).
Fluorescence is a convenient means of observing one molecule at a time, mostly due to the sensitivity of commercial optical detectors, capable of counting single photons.
[6] The MIT team used non-resonance Raman excitation and surface enhancement with silver nanoclusters to detect single cresyl violet molecules, while the team at Indiana University used resonance Raman excitation and surface enhancement with silver nanoparticles to detect single rhodamine 6G molecules.
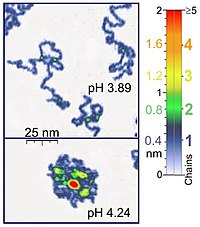
For instance, AFM tapping mode is gentle enough for the recording of adsorbed polyelectrolyte molecules (for example, 0.4 nm thick chains of poly(2-vinylpyridine)) under liquid medium.
At the application of proper scanning parameters, conformation of such molecules remain unchanged for hours that allows the performance of experiments under liquid media having various properties.
Therefore, the functional state of ion channels can be directly measured with sufficiently sensitive electronics, provided that proper precautions are taken to minimize noise.
[9] Single fluorophores can be chemically attached to biomolecules, such as proteins or DNA, and the dynamics of individual molecules can be tracked by monitoring the fluorescent probe.
For instance, single-molecule labeling has yielded a vast quantity of information on how kinesin motor proteins move along microtubule strands in muscle cells.
[12] From the information contained in these unique functions (obtained from individual molecules), one can extract a relatively clear picture on the way the system behaves; e.g. its kinetic scheme, or its potential of activity, or its reduced dimensions form.
For example, it was only after single molecule fluorescence microscopy was used to study kinesin-myosin pairs in muscle tissue that direct observation of the walking mechanisms were understood.
The promise of single molecule in vivo imaging,[16] however, brings with it an enormous potential to directly observe bio-molecules in native processes.