Spatiotemporal gene expression
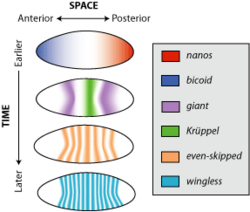
In the early embryonic development of the model organism Drosophila melanogaster, or fruit fly, wingless is expressed across almost the entire embryo in alternating stripes three cells separated.
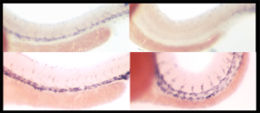
In situ hybridization is an alternate method in which a "probe," a synthetic nucleic acid with a sequence complementary to the mRNA of the gene, is added to the tissue.
A method called enhancer-trap screening reveals the diversity of spatiotemporal gene expression patterns possible in an organism.
Reporter genes can be visualized in living organisms, but both immunohistochemistry and in situ hybridization must be performed in fixed tissues.
Techniques that require fixation of tissue can only generate a single temporal time point per individual organism.
However, using live animals instead of fixed tissue can be crucial in dynamically understanding expression patterns over an individual's lifespan.
To control gene expression spatially inkjet printers are under development for printing ligands on gel culture.
For example, RNA interference can be controlled using light[3][4] and also patterning of gene expression has been performed in cell monolayer[5] and in zebrafish embryos using caged morpholino[6] or peptide nucleic acid[7][8][9] demonstrating the control of gene expression spatiotemporally.