In situ hybridization
In situ hybridization is used to reveal the location of specific nucleic acid sequences on chromosomes or in tissues, a crucial step for understanding the organization, regulation, and function of genes.
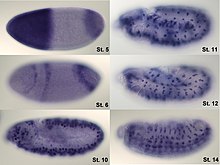
[1][2][3] In situ hybridization is a powerful technique for identifying specific mRNA species within individual cells in tissue sections, providing insights into physiological processes and disease pathogenesis.
A cryostat takes fresh or fixed tissue and immerses it into liquid nitrogen for flash freezing.
[5] For hybridization histochemistry, sample cells and tissues are usually treated to fix the target transcripts in place and to increase access of the probe.
Solution parameters such as temperature, salt, and/or detergent concentration can be manipulated to remove any non-identical interactions (i.e., only exact sequence matches will remain bound).
Then, the probe that was labeled with either radio-, fluorescent- or antigen-labeled bases (e.g., digoxigenin) is localized and quantified in the tissue using either autoradiography, fluorescence microscopy, or immunohistochemistry, respectively.
Next, multiple label probe oligonucleotides (conjugated to alkaline phosphatase or directly to fluorophores) hybridize to each amplifier molecule.
A fully assembled signal amplification structure “Tree” has 400 binding sites for the label probes.