Complementarity (molecular biology)
Complementarity is achieved by distinct interactions between nucleobases: adenine, thymine (uracil in RNA), guanine and cytosine.
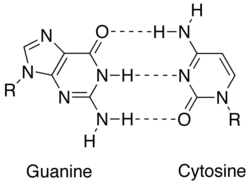
In nucleic acid, nucleobases are held together by hydrogen bonding, which only works efficiently between adenine and thymine and between guanine and cytosine.
[2] Each base pair, A = T vs. G ≡ C, takes up roughly the same space, thereby enabling a twisted DNA double helix formation without any spatial distortions.
This principle plays an important role in DNA replication, setting the foundation of heredity by explaining how genetic information can be passed down to the next generation.
This principle is the basis of commonly performed laboratory techniques such as the polymerase chain reaction, PCR.
Self-complementarity refers to the fact that a sequence of DNA or RNA may fold back on itself, creating a double-strand like structure.
[1] Complementarity can be found between short nucleic acid stretches and a coding region or a transcribed gene, and results in base pairing.
These short nucleic acid sequences are commonly found in nature and have regulatory functions such as gene silencing.
[7] Genome wide studies have shown that RNA antisense transcripts occur commonly within nature.
[8] It has been suggested that complementary regions between sense and antisense transcripts would allow generation of double stranded RNA hybrids, which may play an important role in gene regulation.
[9] miRNAs, microRNA, are short RNA sequences that are complementary to regions of a transcribed gene and have regulatory functions.
And three is by providing a new double-stranded RNA (dsRNA) sequence that Dicer can act upon to create more miRNA to find and degrade more copies of the gene.
Specific characters may be used to create a suitable (ambigraphic) nucleic acid notation for complementary bases (i.e. guanine = b, cytosine = q, adenine = n, and thymine = u), which makes it is possible to complement entire DNA sequences by simply rotating the text "upside down".
[19] For instance, with the previous alphabet, buqn (GTCA) would read as ubnq (TGAC, reverse complement) if turned upside down.
Ambigraphic notations readily visualize complementary nucleic acid stretches such as palindromic sequences.
[20] This feature is enhanced when utilizing custom fonts or symbols rather than ordinary ASCII or even Unicode characters.