Stop codon
[1] Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.
[13] Recognition of stop codons in bacteria have been associated with the so-called 'tripeptide anticodon',[14] a highly conserved amino acid motif in RF1 (PxT) and RF2 (SPF).
[15] Stop codons were historically given many different names, as they each corresponded to a distinct class of mutants that all behaved in a similar manner.
Mutations in viral genes weakened their infectious ability, sometimes creating viruses that were able to infect and grow within only certain varieties of E. coli.
tRNA molecules carrying unnatural aminoacids have been designed to recognize the amber stop codon in bacterial RNA.
Reminiscent of the usual yellow-orange-brown color associated with amber, this second stop codon was given the name of "ochre", an orange-reddish-brown mineral pigment.
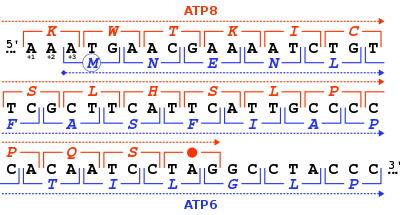
[20] The third and last stop codon in the standard genetic code was discovered soon after, and corresponds to the nucleotide triplet "UGA".
Nonsense mutations are changes in DNA sequence that introduce a premature stop codon, causing any resulting protein to be abnormally shortened.
This often causes a loss of function in the protein, as critical parts of the amino acid chain are no longer assembled.
Most polypeptides resulting from a gene with a nonstop mutation lose their function due to their extreme length and the impact on normal folding.
These prematurely terminate translation if the corresponding frame-shift (such as due to a ribosomal RNA slip) occurs before the hidden stop.
Researchers at Louisiana State University propose the ambush hypothesis, that hidden stops are selected for.
[33] In 2010, when Craig Venter unveiled the first fully functioning, reproducing cell controlled by synthetic DNA he described how his team used frequent stop codons to create watermarks in RNA and DNA to help confirm the results were indeed synthetic (and not contaminated or otherwise), using it to encode authors' names and website addresses.