Reading frame
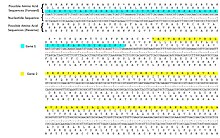
In molecular biology, a reading frame is a way of dividing the sequence of nucleotides in a nucleic acid (DNA or RNA) molecule into a set of consecutive, non-overlapping triplets.
A single strand of a nucleic acid molecule has a phosphoryl end, called the 5′-end, and a hydroxyl or 3′-end.
The codons of the mRNA reading frame are translated in the 5′→3′ direction into amino acids by a ribosome to produce a polypeptide chain.
[5] The usage of multiple reading frames leads to the possibility of overlapping genes; there may be many of these in viral, prokaryote, and mitochondrial genomes.
This is distinct from a frameshift mutation, as the nucleotide sequence (DNA or RNA) is not altered—only the frame in which it is read.
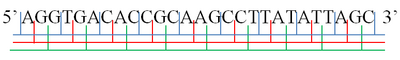
AGG·TGA·CAC·CGC·AAG·CCT·TAT·ATT·AGC
A ·GGT·GAC·ACC·GCA·AGC·CTT·ATA·TTA ·GC
AG ·GTG·ACA·CCG·CAA·GCC·TTA·TAT·TAG· C
A ·GGT·GAC·ACC·GCA·AGC·CTT·ATA·TTA ·GC
AG ·GTG·ACA·CCG·CAA·GCC·TTA·TAT·TAG· C