Mycoplasma laboratorium
Mycoplasma genitalium was originally chosen as the basis for this project because at the time it had the smallest number of genes of all organisms analyzed.
[b 3][a 4] However M. genitalium grows extremely slowly and M. mycoides was chosen as the new focus to accelerate experiments aimed at determining the set of genes actually needed for growth.
[b 4] In 2010, the complete genome of M. mycoides was successfully synthesized from a computer record and transplanted into an existing cell of Mycoplasma capricolum that had had its DNA removed.
[a 5] Mycoplasma is a genus of bacteria of the class Mollicutes in the division Mycoplasmatota (formerly Tenericutes), characterised by the lack of a cell wall (making it Gram negative) due to its parasitic or commensal lifestyle.
In molecular biology, the genus has received much attention, both for being a notoriously difficult-to-eradicate contaminant in mammalian cell cultures (it is immune to beta-lactams and other antibiotics),[a 6] and for its potential uses as a model organism due to its small genome size.
[a 10][a 11] Several newly discovered species have fewer genes than M. genitalium, but are not free-living: many essential genes that are missing in Hodgkinia cicadicola, Sulcia muelleri, Baumannia cicadellinicola (symbionts of cicadas) and Carsonella ruddi (symbiote of hackberry petiole gall psyllid, Pachypsylla venusta[a 12]) may be encoded in the host nucleus.
[a 14][b 6] Several laboratory techniques had to be developed or adapted for the project, since it required synthesis and manipulation of very large pieces of DNA.
Several differences are present in Mycoplasma mycoides JCVI-syn1.0 relative to the reference genome, notably an E.coli transposon IS1 (an infection from the 10kb stage) and an 85bp duplication, as well as elements required for propagation in yeast and residues from restriction sites.
In JCVI-syn1.0 the two species used as donor and recipient are of the same genus, reducing potential problems of mismatches between the proteins in the host cytoplasm and the new genome.
[a 17] Paul Keim (a molecular geneticist at Northern Arizona University in Flagstaff) noted that "there are great challenges ahead before genetic engineers can mix, match, and fully design an organism's genome from scratch".
The 4 watermarks (shown in Figure S1 in the supplementary material of the paper[a 1]) are coded messages written into the DNA, of length 1246, 1081, 1109 and 1222 base pairs respectively.
[a 20] On Oct 6, 2007, Craig Venter announced in an interview with UK's The Guardian newspaper that the same team had synthesized a modified version of the single chromosome of Mycoplasma genitalium chemically.
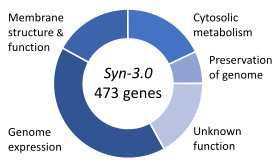
Venter called it “the first designer organism in history” and argued that the fact that 149 of the genes required have unknown functions means that "the entire field of biology has been missing a third of what is essential to life".
[a 21] The project received a large amount of coverage from the press due to Venter's showmanship, to the degree that Jay Keasling, a pioneering synthetic biologist and founder of Amyris commented that "The only regulation we need is of my colleague's mouth".
[b 13] Venter has argued that synthetic bacteria are a step towards creating organisms to manufacture hydrogen and biofuels, and also to absorb carbon dioxide and other greenhouse gases.