Messenger RNA
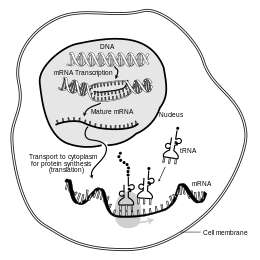
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
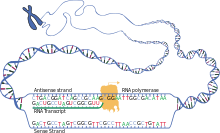
As in DNA, genetic information in mRNA is contained in the sequence of nucleotides, which are arranged into codons consisting of three ribonucleotides each.
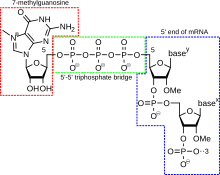
Shortly after the start of transcription, the 5' end of the mRNA being synthesized is bound by a cap-synthesizing complex associated with RNA polymerase.
Although essential for development, the exact role of this editing is not fully understood [7] Polyadenylation is the covalent linkage of a polyadenylyl moiety to a messenger RNA molecule.
In eukaryotic organisms most messenger RNA (mRNA) molecules are polyadenylated at the 3' end, but recent studies have shown that short stretches of uridine (oligouridylation) are also common.
mRNA can also be polyadenylated in prokaryotic organisms, where poly(A) tails act to facilitate, rather than impede, exonucleolytic degradation.
After transcription has been terminated, the mRNA chain is cleaved through the action of an endonuclease complex associated with RNA polymerase.
[9] Mature mRNAs are recognized by their processed modifications and then exported through the nuclear pore by binding to the cap-binding proteins CBP20 and CBP80,[10] as well as the transcription/export complex (TREX).
[21][22] Because prokaryotic mRNA does not need to be processed or transported, translation by the ribosome can begin immediately after the end of transcription.
Translation may occur at ribosomes free-floating in the cytoplasm, or directed to the endoplasmic reticulum by the signal recognition particle.
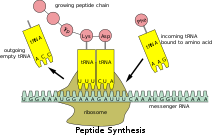
[24][non-primary source needed] Coding regions are composed of codons, which are decoded and translated into proteins by the ribosome; in eukaryotes usually into one and in prokaryotes usually into several.
Genetic variants in 3' UTR have also been implicated in disease susceptibility because of the change in RNA structure and protein translation.
One class of mRNA element, the riboswitches, directly bind small molecules, changing their fold to modify levels of transcription or translation.
An mRNA molecule is said to be monocistronic when it contains the genetic information to translate only a single protein chain (polypeptide).
These polypeptides usually have a related function (they often are the subunits composing a final complex protein) and their coding sequence is grouped and regulated together in a regulatory region, containing a promoter and an operator.
The limited lifetime of mRNA enables a cell to alter protein synthesis rapidly in response to its changing needs.
In some instances, small RNA molecules (sRNA) tens to hundreds of nucleotides long can stimulate the degradation of specific mRNAs by base-pairing with complementary sequences and facilitating ribonuclease cleavage by RNase III.
[37] The poly(A) tail of the mRNA is shortened by specialized exonucleases that are targeted to specific messenger RNAs by a combination of cis-regulatory sequences on the RNA and trans-acting RNA-binding proteins.
Poly(A) tail removal is thought to disrupt the circular structure of the message and destabilize the cap binding complex.
[38] The presence of AU-rich elements in some mammalian mRNAs tends to destabilize those transcripts through the action of cellular proteins that bind these sequences and stimulate poly(A) tail removal.
Detection of a premature stop codon triggers mRNA degradation by 5' decapping, 3' poly(A) tail removal, or endonucleolytic cleavage.
[51] Challenges include the fact that naked RNA sequences naturally degrade after preparation; they may trigger the body's immune system to attack them as an invader; and they are impermeable to the cell membrane.
[49] Overcoming these challenges, mRNA as a therapeutic was first put forward in 1989 "after the development of a broadly applicable in vitro transfection technique.
Gene editing therapies such as CRISPR may also benefit from using mRNA to induce cells to make the desired Cas protein.
[55] The 2023 Nobel Prize in Physiology or Medicine was awarded to Katalin Karikó and Drew Weissman for the development of effective mRNA vaccines against COVID-19.
[60] In 1953, Alfred Hershey, June Dixon, and Martha Chase described a certain cytosine-containing DNA (indicating it was RNA) that disappeared quickly after its synthesis in E.
[62] The idea of mRNA was first conceived by Sydney Brenner and Francis Crick on 15 April 1960 at King's College, Cambridge, while François Jacob was telling them about a recent experiment conducted by Arthur Pardee, himself, and Monod (the so-called PaJaMo experiment, which did not prove mRNA existed but suggested the possibility of its existence).
With Crick's encouragement, Brenner and Jacob immediately set out to test this new hypothesis, and they contacted Matthew Meselson at the California Institute of Technology for assistance.
That fall, Jacob and Monod coined the name "messenger RNA" and developed the first theoretical framework to explain its function.
[62] In February 1961, James Watson revealed that his Harvard-based research group had been right behind them with a series of experiments whose results pointed in roughly the same direction.