Artificial gene synthesis
[3][4] More recently, artificial gene synthesis methods have been developed that will allow the assembly of entire chromosomes and genomes.
The current practical limit is about 200 bp (base pairs) for an oligonucleotide with sufficient quality to be used directly for a biological application.
Usually, a set of individually designed oligonucleotides is made on automated solid-phase synthesizers, purified and then connected by specific annealing and standard ligation or polymerase reactions.
To improve specificity of oligonucleotide annealing, the synthesis step relies on a set of thermostable DNA ligase and polymerase enzymes.
Moreover, because the assembly of the full-length gene product relies on the efficient and specific alignment of long single stranded oligonucleotides, critical parameters for synthesis success include extended sequence regions comprising secondary structures caused by inverted repeats, extraordinary high or low GC-content, or repetitive structures.
Usually these segments of a particular gene can only be synthesized by splitting the procedure into several consecutive steps and a final assembly of shorter sub-sequences, which in turn leads to a significant increase in time and labor needed for its production.
For these annealing based gene synthesis protocols, the quality of the product is directly and exponentially dependent on the correctness of the employed oligonucleotides.
Another problem associated with all current gene synthesis methods is the high frequency of sequence errors because of the usage of chemically synthesized oligonucleotides.
In this case, shorter overlaps do not always allow precise and specific annealing of complementary primers, resulting in the inhibition of full length product formation.
For optimal performance of almost all annealing based methods, the melting temperatures of the overlapping regions are supposed to be similar for all oligonucleotides.
[17] Nevertheless, all these strategies increase time and costs for gene synthesis based on the annealing of chemically synthesized oligonucleotides.
Massively parallel sequencing has also been used as a tool to screen complex oligonucleotide libraries and enable the retrieval of accurate molecules.
[18] In another approach, a complex oligonucleotide library is modified with unique flanking tags before massively parallel sequencing.
Virtually all of the therapeutic proteins in development, such as monoclonal antibodies, are optimised by testing many gene variants for improved function or expression.
In 2012, a group of American scientists led by Floyd Romesberg, a chemical biologist at the Scripps Research Institute in San Diego, California, published that his team designed an unnatural base pair (UBP).
More technically, these artificial nucleotides bearing hydrophobic nucleobases, feature two fused aromatic rings that form a (d5SICS–dNaM) complex or base pair in DNA.
In 2014 the same team from the Scripps Research Institute reported that they synthesized a stretch of circular DNA known as a plasmid containing natural T-A and C-G base pairs along with the best-performing UBP Romesberg's laboratory had designed, and inserted it into cells of the common bacterium E. coli that successfully replicated the unnatural base pairs through multiple generations.
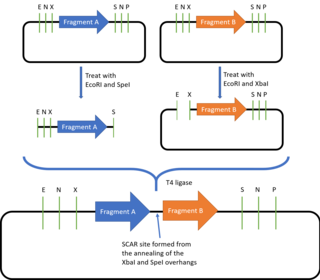
[21] However, because oligonucleotide synthesis typically cannot accurately produce oligonucleotides sequences longer than a few hundred base pairs, DNA assembly methods have to be employed to assemble these parts together to create functional genes, multi-gene circuits or even entire synthetic chromosomes or genomes.
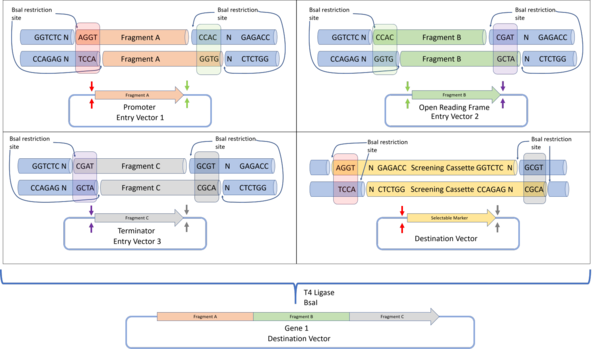
By using the BsaI restriction enzyme that produces a 4 base pair overhang, up to 240 unique, non-palindromic sequences can be used for assembly.
Successfully assembled constructs are selected by detecting the loss of function of a screening cassette that was originally in the destination plasmid.
To enable this construct to be used in a subsequent reaction as an entry vector, the MoClo and Golden Braid standards were designed.
The development of the Golden Gate assembly methods and its variants has allowed researchers to design tool-kits to speed up the synthetic biology workflow.
Instead, integrases make use of unique attachment (att) sites, and catalyse DNA rearrangement between the target fragment and the destination vector.
However, further research revealed that four more orthogonal att sequences could be generated, allowing for the assembly of up to four different DNA fragments, and this process is now known as the Multisite Gateway technology.
These reagents are mixed together with the DNA fragments to be assembled at 50 °C and the following reactions occur: Because the T5 exonuclease is heat labile, it is inactivated at 50 °C after the initial chew back step.
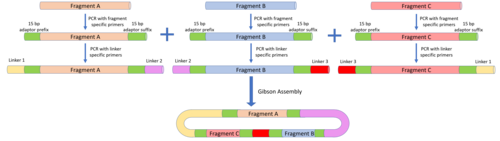
[48] The MODAL strategy defines overlap sequences known as "linkers" to reduce the amount of customisation that needs to be done with each DNA fragment.
To attach these linkers to the parts to be assembled, PCR is carried using part-specific primers containing 15 bp prefix and suffix adaptor sequences.
To allow for idempotent assembly, linkers were also designed with additional methylated iP and iS sequences inserted to protect them from being recognised by BsaI.
[52] On Oct 6, 2007, Craig Venter announced in an interview with UK's The Guardian newspaper that the same team had synthesized a modified version of the single chromosome of Mycoplasma genitalium artificially.
[56] The Yeast 2.0 project applied various DNA assembly methods that have been discussed above, and in March 2014, Jef Boeke of the Langone Medical Centre at New York University, revealed that his team had synthesized chromosome III of S.