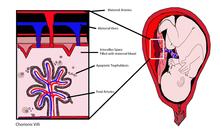
Cell-free fetal DNA
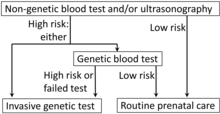
Analysis of cffDNA is a method of non-invasive prenatal diagnosis frequently ordered for pregnant women of advanced age.
As cffDNA is found in maternal blood, sampling carries no associated risk of spontaneous abortion.
[16] New evidence shows that cffDNA test failure rate is higher, fetal fraction (proportion of fetal versus maternal DNA in the maternal blood sample) is lower and PPV for trisomies 18, 13 and SCA is decreased in IVF pregnancies compared to those conceived spontaneously.
Smaller fragments can represent up to seventy percent of the total cell free DNA in the maternal blood sample.
[citation needed] cffDNA may be detected by finding paternally inherited DNA sequences via polymerase chain reaction (PCR).
[27][28] sex-determining region Y gene (SRY) and Y chromosome short tandem repeat "DYS14" in cffDNA from 511 pregnancies were analyzed using quantitative real-time PCR (RT-qPCR).
[30] Microfluidic devices allow the quantification of cffDNA segments in maternal plasma with accuracy beyond that of real-time PCR.
[31][32][33] Digital PCR can differentiate between maternal blood plasma and fetal DNA in a multiplex fashion.
Fetal whole of genome mapping by parental haplotype analysis was completed using sequencing of cffDNA from maternal serum.
[13] Pregnant females were studied using a 2-plex massively parallel maternal plasma DNA sequencing and trisomy was diagnosed with z-score greater than 3.
Then, linear amplification with base extension reaction (with a third primer) is designed to anneal to the region upstream from the mutation site.
When assessing the technique, no false positives or negatives were found when looking for cffDNA to determine fetal sex in sixteen maternal plasma samples.
[39] A technique was described where cffDNA was extracted from maternal plasma and then digested with methylation-sensitive and insensitive restriction enzymes.
For example, Human placental lactogen (hPL) and beta-hCG mRNA are stable in maternal plasma and can be detected.
[42] The main targets in the cffDNA analysis are the gene responsible for the sex-determining region Y protein (SRY) on the Y chromosome and the DYS14 sequence.
[46] Mothers of at risk fetuses are given dexamethasone at 6 weeks gestation to suppress pituitary gland release of androgens.
[47] If analysis of cffDNA obtained from a sample of maternal plasma lacks genetic markers found only on the Y chromosome, it is suggestive of a female fetus.
[citation needed] Autosomal dominant and recessive single gene disorders which have been diagnosed prenatally by analysing paternally inherited DNA include cystic fibrosis, beta thalassemia, sickle cell anemia, spinal muscular atrophy, and myotonic dystrophy.
[50] In studies of the genetics of Huntington's chorea using qRT-PCR of cffDNA from maternal plasma samples, CAG repeats have been detected at normal levels (17, 20 and 24).
[52][32] Incompatibility of fetal and maternal RhD antigens is the main cause of Hemolytic disease of the newborn.
[55] PCR to detect RHD (gene) gene exons 5 and 7 from cffDNA obtained from maternal plasma between 9 and 13 weeks gestation gives a high degree of specificity, sensitivity and diagnostic accuracy (>90 percent) when compared to RhD determination from newborn cord blood serum.
[57] Routine determination of fetal RhD status from cffDNA in maternal serum allows early management of at risk pregnancies while decreasing unnecessary use of Anti-D by over 25 percent.
This trisomy can be detected by analysis of cffDNA from maternal blood by massively parallel shotgun sequencing (MPSS).
Massive parallel sequencing and digital PCR for fetal aneuploidy detection can be used without restriction to fetal-specific nucleic acid molecules.
[76] However, the utility of the procedure may increase as clear associations between specific genetic variants and disease states are discovered.