Common symbiosis signaling pathway
It is known as "common" because different evolutionary younger symbioses also use this pathway, notably the root nodule symbiosis with nitrogen-fixing rhizobia bacteria.
A recent work [1] by Kevin Cope and colleagues showed that ectomycorrhizae (a different type of mycorrhizae) also uses CSSP components such as Myc-factor recognition.
The AMF colonization requires the following chain[2] of events that can be roughly divided into the following steps: 1: Pre-Contact Signaling
2: C: Transcription3: The Accommodation program To accurately recognize the infection thread of a different species of organism, and to establish a mutually beneficial association requires robust signaling.
[5] At the pre-symbiotic signaling, plants and AMF release chemical factors in their surroundings so that the partners can recognise and find each other.
[6]' Plant root exudates play roles in complex microbial interaction,[7] by releasing a variety of compounds,[7][8][9] among which strigolactone has been identified to facilitate both AMF colonisation and pathogen infection.
The chemical nature of some Myc-factors has recently been revealed as lipo-chito-oligosaccharide (Myc-LCOs) and chito-oligosaccharides (Myc-COs) that work as symbiotic signal.
[2] CCaMK and CYCLOPS probably forms a complex that along with DELLA protein, regulates the gene expression of RAM1 (Reduced Arbuscular Mycorrhyza1).
This includes invagination of host plasmalemma, proliferation of endoplasmic reticulum, golgi apparatus, trans-golgi network and secretory vesicles.
From the host plant's side, it synthesizes and releases a range of caroteinoid based phytohormone, called strigolactones.
[2] NOPE1 or 'NO PERCEPTION 1', is a transporter protein in Rice (Oryza sativa) and Maize (Zea mays), also required for the priming stage for colonisation by the fungus.
In Lotus japonicus, LYS11, a receptor for LCOs, was expressed in root cortex cells associated with intra-radical colonizing arbuscular mycorrhizal fungi [24] AM host plants show symbiotic-activated calcium waves upon exposure to short chain chitin oligomers.
The lysine motif domain of OsCERK1 and OsCEBiP is thought to be involved in the perception of short chain chitin oligomers.
NOPE-1 also shows a strong N-acetylglucosamine uptake activity, and is thought to be associated with recognition of presence of fungal symbiont.
[2] Some plant proteins are suspected to recognise Myc-factors, and the rice OsCERK1 Lysin motif (LysM) receptor-like kinase, is one of them.
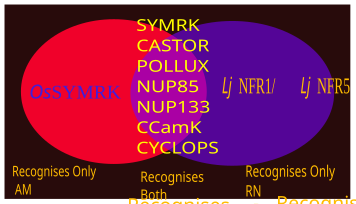
[25][26][27][28] DMI2/ SYMRK is a receptor-like kinase, an important protein in endosymbiosis signal perception, reported in several plants (Mt-DMI2 or Mt-NORK in Medicago trancatula; Lj-SYMRK in Lotus japonicas; Ps-SYM19 in Pisum sativum; OsSYMRK in Rice).
[29] Notably, it has been found that a Nod-factor inducible gene, MtENOD11, is activated in the presence of AMF exudates; little is known about this phenomenon.
Tomato (Solanum lycopersicum), a non-legume eudicot, also have a similar LysM receptor, SlLYK10 that Promotes AM symbiosis.
[35] Some elements involved in this process are: Lotus japonicus Nucleoporins LjNUP85 and LjNUP133 has potential role in transmission of the signal.
[43] Root cortex cells experience important changes in order to accommodate for the fungal endosymbiont.
[44] Although various proteins have been identified which may play role on how this accommodation process occurs, the detailed signalling cascade is not fully understood.
[37] Plant enzymes FatM and RAM2[45] and ABC transporter STR/STR2 are putatively involved in the synthesis and supplying of a lipid 16:0 β-monoacylglycerol to the AM fungi.