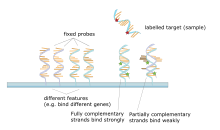DNA microarray
Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome.
An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis.
A high number of complementary base pairs in a nucleotide sequence means tighter non-covalent bonding between the two strands.
[10] Microarrays can be manufactured in different ways, depending on the number of probes under examination, costs, customization requirements, and the type of scientific question being asked.
In spotted microarrays, the probes are oligonucleotides, cDNA or small fragments of PCR products that correspond to mRNAs.
This provides a relatively low-cost microarray that may be customized for each study, and avoids the costs of purchasing often more expensive commercial arrays that may represent vast numbers of genes that are not of interest to the investigator.
Publications exist which indicate in-house spotted microarrays may not provide the same level of sensitivity compared to commercial oligonucleotide arrays,[13] possibly owing to the small batch sizes and reduced printing efficiencies when compared to industrial manufactures of oligo arrays.
More recently, Maskless Array Synthesis from NimbleGen Systems has combined flexibility with large numbers of probes.
Each RNA molecule encounters protocol and batch-specific bias during amplification, labeling, and hybridization phases of the experiment making comparisons between genes for the same microarray uninformative.
Another benefit is that data are more easily compared to arrays from different experiments as long as batch effects have been accounted for.
Third, spots of each cDNA clone or oligonucleotide are present as replicates (at least duplicates) on the microarray slide, to provide a measure of technical precision in each hybridization.
It is critical that information about the sample preparation and handling is discussed, in order to help identify the independent units in the experiment and to avoid inflated estimates of statistical significance.
[20] Microarray data is difficult to exchange due to the lack of standardization in platform fabrication, assay protocols, and analysis methods.
Various grass-roots open-source projects are trying to ease the exchange and analysis of data produced with non-proprietary chips: For example, the "Minimum Information About a Microarray Experiment" (MIAME) checklist helps define the level of detail that should exist and is being adopted by many journals as a requirement for the submission of papers incorporating microarray results.
[21] The MGED Society has developed standards for the representation of gene expression experiment results and relevant annotations.
[33] Other methods permit analysis of data consisting of a low number of biological or technical replicates; for example, the Local Pooled Error (LPE) test pools standard deviations of genes with similar expression levels in an effort to compensate for insufficient replication.
A number of open-source data warehousing solutions, such as InterMine and BioMart, have been created for the specific purpose of integrating diverse biological datasets, and also support analysis.
Advances in massively parallel sequencing has led to the development of RNA-Seq technology, that enables a whole transcriptome shotgun approach to characterize and quantify gene expression.