Deep homology
[1] In 1978, Edward B. Lewis helped to found evolutionary developmental biology, discovering that homeotic genes regulated embryonic development in fruit flies.
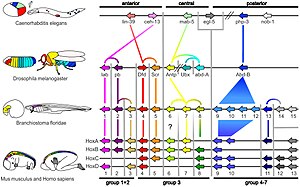
[1] In 1997, the term deep homology first appeared in a paper by Neil Shubin, Cliff Tabin, and Sean B. Carroll, describing the apparent relatedness in genetic regulatory apparatuses which indicated evolutionary similarities in disparate animal features.
HoxA and HoxD, that regulate finger and toe formation in mice, control the development of ray fins in zebrafish; these structures had until then been considered non-homologous.
[7] In modern day biology, the depth of understanding deep homology has evolved into focusing on the molecular and genetic mechanisms and functions rather than simple morphology.
[9] In 2010, a team led by Edward Marcotte developed an algorithm that identifies deeply homologous genetic modules in unicellular organisms, plants, and animals based on phenotypes (such as traits and developmental defects).