Elementary modes
In biochemistry, elementary modes[1] may be considered minimal realizable flow patterns through a biochemical network that can sustain a steady state.
This means that elementary modes cannot be decomposed further into simpler pathways.
All possible flows through a network can be constructed from linear combinations of the elementary modes.
The set of elementary modes for a given network is unique (up to an arbitrary scaling factor).
the vector of steady state floating (or internal) species and
An important condition is that the rate of each irreversible reaction must be non-negative,
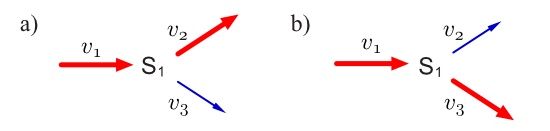
Consider a simple branched pathway with all three steps irreversible.
Such a pathway will admit two elementary modes which are indicated in thicked (or red) reaction lines.
For condition two we must ensure that all reactions that are irreversible have positive entries in the corresponding elements of the elementary modes.
Since all three reactions in the branch are irreversible and all entries in the elementary modes are positive, condition two is satisfied.
Finally, to satisfy condition three, we must ask whether we can decompose the two elementary modes into other paths that can sustain a steady state while using the same non-zero entries in the elementary mode.
In this example, it is impossible to decompose the elementary modes any further without disrupting the ability to sustain a steady state.
Therefore, with all three conditions satisfied, we can conclude that the two vectors shown above are elementary modes.
All possible flows through a network can be constructed from linear combinations of the elementary modes, that is:
must be greater than or equal to zero to ensure that irreversible steps aren't inadvertently made to go in the reverse direction.
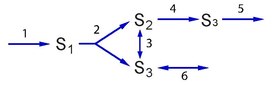
If one of the outflow steps in the simple branched pathway is made reversible, an additional elementary mode becomes available, representing the flow between the two outflow branches.
This means that any sequence of reactions can be labeled a metabolic pathway.
However, as metabolism was being uncovered, groups of reactions were assigned specific labels, such as glycolysis, Krebs Cycle, or Serine biosynthesis.
Often the categorization was based on common chemistry or identification of an input and output.
This is a somewhat ad hoc means for defining pathways, particularly when pathways are dynamic structures, changing as environmental result in changes in gene expression.
In E. coli and other bacteria, it is only cyclic during aerobic growth on acetate or fatty acids.
[3] Instead, under anaerobiosis, its enzymes function as two distinct biosynthetic pathways producing succinyl-CoA and α-ketoglutarate.
The added advantage is that the set of elementary modes is unique and non-decomposable to simpler pathways.
Elementary modes, therefore, provide an unambiguous definition of a pathway.
The two other important features as indicated before are pathway uniqueness and thermodynamic plausibility.
That is, is there more than one way to generate a pathway (i.e., something that can sustain a steady state) with the enzymes currently used in the mode?
To illustrate this subtle condition, consider the pathway shown in below.
Step three and six are reversible and correspond to triose phosphate isomerase and glycerol 3-phosphate dehydrogenase, respectively.
The network has four elementary flux modes, which are shown in the figure below.
Note that it is possible to have negative entries in the set of elementary modes because they will correspond to the reversible steps.