Fluorescence correlation spectroscopy
[1] In contrast to other methods (such as HPLC analysis) FCS has no physical separation process; instead, it achieves its spatial resolution through its optics.
In these techniques light is focused on a sample and the measured fluorescence intensity fluctuations (due to diffusion, physical or chemical reactions, aggregation, etc.)
When an appropriate model is known, FCS can be used to obtain quantitative information such as Because fluorescent markers come in a variety of colors and can be specifically bound to a particular molecule (e.g. proteins, polymers, metal-complexes, etc.
With the development of sensitive detectors such as avalanche photodiodes the detection of the fluorescence signal coming from individual molecules in highly dilute samples has become practical.
With this emerged the possibility to conduct FCS experiments in a wide variety of specimens, ranging from materials science to biology.
[4] Signal-correlation techniques were first experimentally applied to fluorescence in 1972 by Magde, Elson, and Webb,[5] who are therefore commonly credited as the inventors of FCS.
[9] The former led to an analysis of distributions and moments of the fluorescent signals for extracting molecular information,[10][11] which eventually became a collection of methods known as Brightness Analyses.
[17] The measurement volume is a convolution of illumination (excitation) and detection geometries, which result from the optical elements involved.
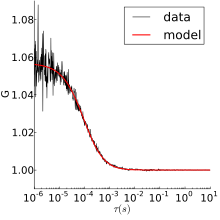
[18] One common way of calibrating the measurement volume parameters is to perform FCS on a species with known diffusion coefficient and concentration (see below).
the dynamics is often not sufficiently well-described by the normal diffusion model, where the mean squared displacement (MSD) grows linearly with time.
Also, a power law is, in a strict sense, the expected form only for a narrow range of rigorously defined systems, for instance when the distribution of obstacles is fractal.
A wide range of possible FCS experiments involve chemical reactions that continually fluctuate from equilibrium because of thermal motions (and then "relax").
One very simple system showing chemical relaxation would be a stationary binding site in the measurement volume, where particles only produce signal when bound (e.g. by FRET, or if the diffusion time is much faster than the sampling interval).
The following table gives diffusion coefficients of some common fluorophores in water at room temperature, and their excitation wavelengths.
The relationship between the diffusion time and the spot area is linear and could be plotted in order to decipher the major contribution of confinement.
[47] Fluorescence cross correlation spectroscopy overcomes the weak dependence of diffusion rate on molecular mass by looking at multicolor coincidence.
These methods use the heterogeneity in the intensity distribution of fluorescence to measure the molecular brightness of different species in a sample.
The brightness analysis method might be employed to study the interactions of biomolecules upon binding a non-fluorescent reactant to a fluorescent one.
[49] The complex formation causes a change in brightness intensity due to steric shielding, charge transfer, photoisomerization rate, or a combination of these phenomena enabling distinguishing the reactant from the product.
In Scanning fluorescence correlation spectroscopy (sFCS) the measurement volume is moved across the sample in a defined way.
The introduction of scanning is motivated by its ability to alleviate or remove several distinct problems often encountered in standard FCS, and thus, to extend the range of applicability of fluorescence correlation methods in biological systems.
Raster ICS (RICS),[52] and position sensitive FCS (PSFCS)[53] incorporate the time delay between parts of the image scan into the analysis.
[59] When the motion is slow (in biology, for example, diffusion in a membrane), getting adequate statistics from a single-point FCS experiment may take a prohibitively long time.
More data can be obtained by performing the experiment in multiple spatial points in parallel, using a laser scanning confocal microscope.
A variation that is closely related to STICS (by the Fourier transform) is k-space Image Correlation Spectroscopy (kICS).
Adapted from methods of spatio-temporal image correlation spectroscopy,[59] it exploits the high positional accuracy of single-particle tracking.
A particle image cross-correlation spectroscopy (PICCS) extension is available for biological processes that involve multiple interaction partners, as can observed by two-color microscopy.
[68] Because the fluorescence intensity in TIRF falls off exponentially with distance from the coverslip (instead of as a Gaussian with a confocal), the autocorrelation function is different.
FCS experiments require a level of processing and are more sensitive to potentially confounding influences like: rotational diffusion, vibrations, photobleaching, dependence on illumination and fluorescence color, inadequate statistics, etc.
[71] [72] [73] Several advantages in both spatial resolution and minimizing photodamage/photobleaching in organic and/or biological samples are obtained by two-photon or three-photon excitation FCS.