Frameshift mutation
Frameshift mutations have been proposed as a source of biological novelty, as with the alleged creation of nylonase, however, this interpretation is controversial.
A study by Negoro et al. (2006)[4] found that a frameshift mutation was unlikely to have been the cause and that rather a two amino acid substitution in the active site of an ancestral esterase resulted in nylonase.
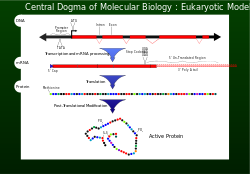
In 1956 Francis Crick described the flow of genetic information from DNA to a specific amino acid arrangement for making a protein as the central dogma.
An incorrectly made protein can have detrimental effects on cell viability and in most cases cause the higher organism to become unhealthy by abnormal cellular functions.
To ensure that the genome successfully passes the information on, proofreading mechanisms such as exonucleases and mismatch repair systems are incorporated in DNA replication.
Reverse transcriptase, as opposed to RNA Polymerase II, is thought to be a stronger cause of the occurrence of frameshift mutations.
Guide RNA can also be used to insert or delete Uridine into the mRNA after transcription, this allows for the correct reading frame.
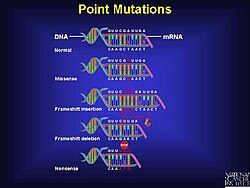
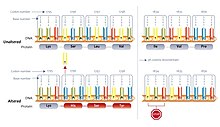
For instance, the addition of an extra "A" leads to a sequence shift, triggering the reading of an entirely different set of codons.
In most instances, the new reading frame results in an early encounter with a stop codon, leading to the formation of a shortened and usually inactive protein.
An environmental study, specifically the production of UV-induced frameshift mutations by DNA polymerases deficient in 3′ → 5′ exonuclease activity was done.
[7] The effects of neighboring bases and secondary structure to detect the frequency of frameshift mutations has been investigated in depth using fluorescence.
[8] Studies on the effects of the length of the primer strand reveal that an equilibrium mixture of four hybridization conformations was observed when template bases looped-out as a bulge, i.e. a structure flanked on both sides by duplex DNA.
[9] Sanger sequencing and pyrosequencing are two methods that have been used to detect frameshift mutations, however, it is likely that data generated will not be of the highest quality.
[12] An experiment to determine the accuracy of this newer sequencing method tested for 21 genes and had no false positive calls for frameshift mutations.
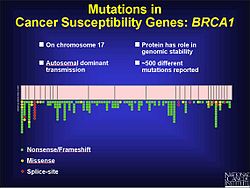
[1] Small insertions or deletions (those less than 20 base pairs) make up 24% of mutations that manifest in currently recognized genetic disease.
[15] Experiments can be run to determine the frequency of the frameshift mutation by adding or removing a pre-set number of nucleotides.
[15] Huntington's disease is one of the nine codon reiteration disorders caused by polyglutamine expansion mutations that include spino-cerebellar ataxia (SCA) 1, 2, 6, 7 and 3, spinobulbar muscular atrophy and dentatorubal-pallidoluysianatrophy.
There may be a link between diseases caused by polyglutamine and polyalanine expansion mutations, as frame shifting of the original SCA3 gene product encoding CAG/polyglutamines to GCA/polyalanines.
Currently there are attempts to use frameshift mutations beneficially in the treatment of diseases, changing the reading frame of the amino acids.
Experiments in yeast and bacteria help to show characteristics of microsatellites that may contribute to defective DNA mismatch repair.
[17] In prostate cancer a frameshift mutation changes the open reading frame (ORF) and prevents apoptosis from occurring.
This region on the open reading frame ORF contains a frameshift mutation leading to a premature stop codon.
A 4 base pair insertion in exon 11 is observed in 80% of Tay-Sachs disease presence in the Ashkenazi Jewish population.
The majority of SMS cases harbor an ~3.5 Mb common deletion that encompasses the retinoic acid induced-1 (RAI1) gene.
Other cases illustrate variability in the SMS phenotype not previously shown for RAI1 mutation, including hearing loss, self-abusive behaviours, and mild global delays.
The results indicate that this heptameric C-tract is a preferential recombination hotspot insertion/deletions (SNindels) and therefore a primary target for analysis in patients suspected for mutations in RAI1.
A recent study has indicated that a frameshift mutation (c.363dupG or p.Gln122AlafsX30) in Troponin C was the cause of hypertrophic cardiomyopathy (and sudden cardiac death) in a 19-year-old male.
Gene therapy procedures include modifying the zinc fringer nuclease fusion protein, cleaving both ends of the mutation, which in turn removes it from the sequence.
This process allows for passing over the mutation so that the rest of the sequence remains in frame and the function of the protein stays intact.
The idea is to use immunotherapy with combinatorial mixtures of tumour-specific frameshift mutation-derived peptides to elicit a cytotoxic T-cell response specifically directed against tumour cells.