GENCODE
[9] The pilot stage of the ENCODE project aimed to investigate in great depth, computationally and experimentally, 44 regions totaling 30 Mb of sequence representing approximately 1% of the human genome.
[10] The findings highlighted the success of the pilot project to create a feasible platform and new technologies to characterise functional elements in the human genome, which paves the way for opening research into genome-wide studies.
2007 October New funding was part of NHGRI's endeavour to scale-up the ENCODE Project to a production phase on the entire genome along with additional pilot-scale studies.
The new track was designed to assist in the search for appropriate guide sentences by listing potential binding sites for the CRISPR/Cas9 complex that are next to transcribed regions, or within 200 bp of one.
Also, given the COVID-19 pandemic during 2020, there has been an urge to support research responding to the situation, so GENCODE has reviewed and improved the annotation for a set of protein-coding genes associated with SARSCoV-2 infection.
A summary of key participating institutions of each phase is listed below:[6][13] Source:[8] Since its inception, GENCODE has released 36 versions of the Human gene set annotations (excluding minor updates).
This is mostly attributed to new experimental evidence obtained using Cap Analysis Gene Expression (CAGE) clusters, annotated PolyA sites, and peptide hits.
All Ensembl transcripts are based on experimental evidence and thus the automated pipeline relies on the mRNAs and protein sequences deposited into public databases from the scientific community.
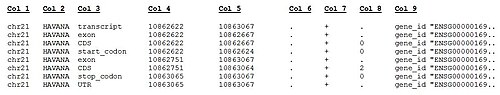
TAB-separated standard GTF columns Description of key-value pairs in 9th column of the GENCODE GTF file (format: key "value") Also, the GENCODE website contains a Genome Browser for human and mouse where you can reach any genomic region by giving the chromosome number and start-end position (e.g. 22:30,700,000..30,900,000), as well as by ENS transcript id (with/without version), ENS gene id (with/without version) and gene name.
First, genes were conceived in the 1900s as discrete units of heredity, then it was thought as the blueprint for protein synthesis, and in more recent times, it was being defined as genetic code that is transcribed into RNA.
With the advent of the ENCODE/GENCODE project, even more problematic aspects of the definition have been uncovered, including alternative splicing (where a series of exons are separated by introns), intergenic transcriptions, and the complex patterns of dispersed regulation, together with non-genic conservation and the abundance of noncoding RNA genes.
[22] A key research area of the GENCODE project was to investigate the biological significance of long non-coding RNAs (lncRNA).
The primary goals of RGASP are to provide an unbiased evaluation for RNA-seq alignment, transcript characterisation (discovery, reconstruction and quantification) software, and to determine the feasibility of automated genome annotations based on transcriptome sequencing.
One of the main discoveries from rounds 1 & 2 of the project was the importance of read alignment on the quality of gene predictions produced.