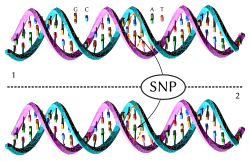
Genealogical DNA test
However, due to the random nature of which, and how much, DNA is inherited by each tested person from their common ancestors, precise relationship conclusions can only be made for close relations.
Typically, the sample collection uses a home test kit supplied by a service provider such as 23andMe, AncestryDNA, Family Tree DNA, or MyHeritage.
[8] The preparation of a report on the DNA in the sample proceeds in multiple stages: All major service providers use equipment with chips supplied by Illumina.
Raw data can also be uploaded to services that provide health risk and trait reports using SNP genotypes.
[16] The X-chromosome has a special path of inheritance patterns and can be useful in significantly narrowing down possible ancestor lines compared to autosomal DNA.
The number of STRs offered is limited, and results have been used for personal identification,[20] paternity cases, and inter-population studies.
A perfect match found to another person's mtDNA test results indicates shared ancestry of possibly between 1 and 50 generations ago.
Generally, testing only the HVRs has limited genealogical use so it is increasingly popular and accessible to have a full sequence.
The full mtDNA sequence is only offered by Family Tree DNA among the major testing companies[26] and is somewhat controversial because the coding region DNA may reveal medical information about the test-taker[27] All humans descend in the direct female line from Mitochondrial Eve, a female who lived probably around 150,000 years ago in Africa.
Instead, results are normally compared to the Cambridge Reference Sequence (CRS), which is the mitochondria of a European who was the first person to have their mtDNA published in 1981 (and revised in 1999).
DNA companies will usually provide an estimate of how closely related two people are, in terms of generations or years, based on the difference between their results.
A true genetic relationship can only be determined using Y-DNA SNP mutations, but sequencing these was historically more time-consuming and expensive.
Unique permanent SNP mutations occur in every male line every 83 years on average,[35] providing excellent time resolution.
All human men descend in the paternal line from a single man dubbed Y-chromosomal Adam, who lived probably between 200,000 and 300,000 years ago.
For recent genealogy, exact matching on the mtDNA full sequence is used to confirm a common ancestor on the direct maternal line between two suspected relatives.
Geneticist Adam Rutherford has written that these tests "don’t necessarily show your geographical origins in the past.
African Americans usually cannot easily trace their ancestry during the years of slavery through surname research, census and property records, and other traditional means.
The historical research of Paul Heinegg has documented that many of the Melungeon groups in the Upper South were descended from mixed-race people who were free in colonial Virginia and the result of unions between the Europeans and Africans.
They moved to the frontiers of Virginia, North Carolina, Kentucky and Tennessee to gain some freedom from the racial barriers of the plantation areas.
[46] Several efforts, including a number of ongoing studies, have examined the genetic makeup of families historically identified as Melungeon.
[48] Native American tribes have their own requirements for membership, often based on at least one of a person's ancestors having been included on tribal-specific Native American censuses (or final rolls) prepared during treaty-making, relocation to reservations or apportionment of land in the late 19th century and early 20th century.
This could be consistent with a shared common ancestor, or with the hereditary priesthood having originally been founded from members of a single closely related clan.
A further academic study published in 2009 examined more STR markers and identified a more sharply defined SNP haplogroup, J1e* (now J1c3, also called J-P58*) for the J1 lineage.
The research found "that 46.1% of Kohanim carry Y chromosomes belonging to a single paternal lineage (J-P58*) that likely originated in the Near East well before the dispersal of Jewish groups in the Diaspora.
Y-DNA testing results are normally stated as probabilities: For example, with the same surname a perfect 37/37 marker test match gives a 95% likelihood of the most recent common ancestor (MRCA) being within 8 generations,[51] while a 111 of 111 marker match gives the same 95% likelihood of the MRCA being within only 5 generations back.
[81] Promethease, and its research paper crawling database SNPedia, has received criticism for technical complexity and a poorly defined "magnitude" scale that causes misconceptions, confusion and panic among its users.
In fall 2001, GeneTree sold its assets to Salt Lake City-based Sorenson Molecular Genealogy Foundation (SMGF) which originated in 1999.
[88] Later, GeneTree returned to genetic testing for genealogy in conjunction with the Sorenson parent company and eventually was part of the assets acquired in the Ancestry.com buyout of SMGF in 2012.
[89][90] In 2000, Family Tree DNA, founded by Bennett Greenspan and Max Blankfeld, was the first company dedicated to direct-to-consumer testing for genealogy research.
[97][98] MyHeritage launched its genetic testing service in 2016, allowing users to use cheek swabs to collect samples.