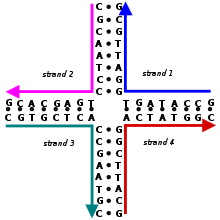
Holliday junction
Holliday junctions may exist in a variety of conformational isomers with different patterns of coaxial stacking between the four double-helical arms.
Coaxial stacking is the tendency of nucleic acid blunt ends to bind to each other, by interactions between the exposed bases.
The unstacked form dominates in the absence of divalent cations such as Mg2+, because of electrostatic repulsion between the negatively charged backbones of the strands.
As of 2000, it was not known with certainty whether the electrostatic shielding was the result of site-specific binding of cations to the junction, or the presence of a diffuse collection of the ions in solution.
[1] In addition, cruciform structures involving Holliday junctions can arise to relieve helical strain in symmetrical sequences in DNA supercoils.
Other classes are branch migration proteins that increase the exchange rate by orders of magnitude, and site-specific recombinases.
[1] In prokaryotes, Holliday junction resolvases fall into two families, integrases and nucleases, that are each structurally similar although their sequences are not conserved.
[8] In bacteria, branch migration is facilitated by the RuvABC complex or RecG protein, molecular motors that use the energy of ATP hydrolysis to move the junction.
[12] The pathway that produces the majority of crossovers in S. cerevisiae budding yeast, and possibly in mammals, involves proteins EXO1, MLH1-MLH3 heterodimer (called MutL gamma) and SGS1 (ortholog of Bloom syndrome helicase).
[15] While the other three pathways, involving proteins MUS81-MMS4, SLX1 and YEN1, respectively, can promote Holliday junction resolution in vivo, absence of all three nucleases has only a modest impact on formation of crossover products.
[20] Nevertheless, this mutant gave rise to spore viability patterns suggesting that segregation of non-exchange chromosomes occurred efficiently.
[23][24] The most common such motif is the double crossover (DX) complex, which contains two Holliday junctions in close proximity to each other, resulting in a rigid structure that can self-assemble into larger arrays.
Tiles of three Holliday junctions in a triangular fashion have been used to make periodic three-dimensional arrays for use in X-ray crystallography of biomolecules.
[23][24] Robin Holliday proposed the junction structure that now bears his name as part of his model of homologous recombination in 1964, based on his research on the organisms Ustilago maydis and Saccharomyces cerevisiae.
Holliday realized that the proposed pathway would create heteroduplex DNA segments with base mismatches between different versions of a single gene.
[28] In the original Holliday model for homologous recombination, single-strand breaks occur at the same point on one strand of each parental DNA.
Depending on which strand was used as a template to repair the other, the four cells resulting from meiosis might end up with three copies of one allele and only one of the other, instead of the normal two of each, a property known as gene conversion.
[3] Holliday's original model assumed that heteroduplex DNA would be present on both chromosomes, but experimental data on yeast refuted this.
In 1983, artificial Holliday junction molecules were first constructed from synthetic oligonucleotides by Nadrian Seeman, allowing for more direct study of their physical properties.
Much of the early analysis of Holliday junction structure was inferred from gel electrophoresis, FRET, and hydroxyl radical and nuclease footprinting studies.
[1][3][29] Initially, geneticists assumed that the junction would adopt a parallel rather than antiparallel conformation, because that would place the homologous duplexes in closer alignment to each other.
[24][31] Seeman developed the more rigid double-crossover (DX) motif, suitable for forming two-dimensional lattices, demonstrated in 1998 by him and Erik Winfree.
[32] The synthesis of a three-dimensional lattice was finally published by Seeman in 2009, nearly thirty years after he had set out to achieve it.