Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids.
Wilcox, and T.J. Kelly, working at Johns Hopkins University in 1968, isolated and characterized the first restriction nuclease whose functioning depended on a specific DNA nucleotide sequence.
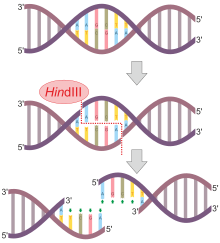
Working with Haemophilus influenzae bacteria, this group isolated an enzyme, called HindII, that always cut DNA molecules at a particular point within a specific sequence of six base pairs.
Most nucleases are classified by the Enzyme Commission number of the "Nomenclature Committee of the International Union of Biochemistry and Molecular Biology" as hydrolases (EC-number 3).
In the case of endonucleases such as EcoRV, BamHI, and PvuII, this nonspecific binding involves electrostatic interactions between minimal surface area of the protein and the DNA.
This results in significant deformation of the DNA tertiary structure and is accomplished with a surfaces rich in basic (positively charged) residues.
However most are nonspecific, instead recognizing structural abnormalities produced in the DNA backbone by base pair mismatches.
There are more than 900 restriction enzymes, some sequence specific and some not, have been isolated from over 230 strains of bacteria since the initial discovery of HindII.
Numbers following the nuclease names indicate the order in which the enzymes were isolated from single strains of bacteria: EcoRI, EcoRII.
Once it encounters its particular specific recognition sequence, it will bind to the DNA molecule and makes one cut in each of the two sugar-phosphate backbones.
Once the cuts have been made, the resulting fragments are held together only by the relatively weak hydrogen bonds that hold the complementary bases to each other.
They recognize damage sites through deformation of double stranded DNA (dsDNA) secondary structure.
Deletions inactivating or removing these nucleases increase rates of mutation and mortality in affected microbes and cancer in mice.
The MutSLH system (comprising MutS, MutL, and MutH) corrects point mutations and small turns.
One of the exonucleases RecJ, ExoVII, or ExoI then degrades the site before DNA polymerase resynthesizes the gap in the strand.
It is the result of spontaneous hydrolysis and the activity of DNA glycosylases as an intermediary step in base excision repair.
Instances of crosslinking, adducts, and lesions (generated by ultraviolet light or reactive oxygen species) can trigger this repair pathway.
Deletions or mutations which affect these nucleases instigate increased sensitivity to ultraviolet damage and carcinogenesis.
Unintentional breaks are commonly generated by ionizing radiation, various exogenous and endogenous chemical agents, and halted replication forks.
Both cases require the ends in double strand breaks be processed by nucleases before repair can take place.
[9] V(D)J recombination involves opening stem-loops structures associated with double-strand breaks and subsequently joining both ends.
[9] The frequency at which a particular nuclease will cut a given DNA molecule depends on the complexity of the DNA and the length of the nuclease's recognition sequence; due to the statistical likelihood of finding the bases in a particular order by chance, a longer recognition sequence will result in less frequent digestion.
One unique family of nucleases is the meganucleases, which are characterized by having larger, and therefore less common, recognition sequences consisting of 12 to 40 base pairs.
These nucleases are particularly useful for genetic engineering and Genome engineering applications in complex organisms such as plants and mammals, where typically larger genomes (numbering in the billions of base pairs) would result in frequent and deleterious site-specific digestion using traditional nucleases.