Human evolutionary genetics
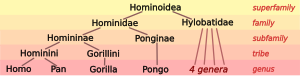
Distance on a phylogenetic tree can be used approximately to indicate: The separation of humans from their closest relatives, the non-human African apes (chimpanzees and gorillas), has been studied extensively for more than a century.
The gorilla was, in ID terms, closer to human than to chimpanzees; however, the difference was so slight that the trichotomy could not be resolved with certainty.
Later studies based on molecular genetics were able to resolve the trichotomy: chimpanzees are phylogenetically closer to humans than to gorillas.
Usually the molecular clock is calibrated assuming that the orangutan split from the African apes (including humans) 12-16 MYA.
If true that means that the human lineage would have experienced an immense decrease of its effective population size (and thus genetic diversity) in its evolution.
(see Toba catastrophe theory) Another study (Chen & Li, 2001) sequenced 53 non-repetitive, intergenic DNA segments from human, chimpanzee, gorilla and orangutan.
Therefore, if the internodal time span is known, the ancestral effective population size of the common ancestor of humans and chimpanzees can be calculated.
The internodal time span is useful to estimate the ancestral effective population size of the common ancestor of humans and chimpanzees.
This value is not as high as that from the first study (Takahata), but still much higher than present day effective population size of humans.
A third study (Yang, 2002) used the same dataset that Chen and Li used but estimated the ancestral effective population of 'only' ~12,000 to 21,000, using a different statistical method.
Based on the assumption of a constant molecular clock, the study predicts the gene loss occurred relatively recently in human evolution—less than 240 000 years ago, but both the Vindija Neandertal and the high-coverage Denisovan sequence contain the same premature stop codons as modern humans and hence dating should be greater than 750 000 years ago.
[20] Stedman et al. (2004) stated that the loss of the sarcomeric myosin gene MYH16 in the human lineage led to smaller masticatory muscles.
They estimated that the mutation that led to the inactivation (a two base pair deletion) occurred 2.4 million years ago, predating the appearance of Homo ergaster/erectus in Africa.
The period that followed was marked by a strong increase in cranial capacity, promoting speculation that the loss of the gene may have removed an evolutionary constraint on brain size in the genus Homo.
[22] Segmental duplications (SDs or LCRs) have had roles in creating new primate genes and shaping human genetic variation.
These regions show signs of being subject to natural selection, leading to the evolution of distinctly human traits.
Analyses of conserved non-coding sequences, which often contain functional and thus positively selected regulatory regions, address this possibility.
Of this, 1.06% or less was thought to represent fixed differences between the species, with the rest being variant sites in humans or chimpanzees.
The sequence divergence has generally the following pattern: Human-Chimp < Human-Gorilla << Human-Orangutan, highlighting the close kinship between humans and the African apes.
Alu elements diverge quickly due to their high frequency of CpG dinucleotides which mutate roughly 10 times more often than the average nucleotide in the genome.
[16] The measures of sequence divergence shown in the table only take the substitutional differences, for example from an A (adenine) to a G (guanine), into account.
Neanderthals and most modern humans share a lactose-intolerant variant of the lactase gene that encodes an enzyme that is unable to break down lactose in milk after weaning.
[30][31] With their rapid expansion throughout different climate zones, and especially with the availability of new food sources with the domestication of cattle and the development of agriculture, human populations have been exposed to significant selective pressures since their dispersal.
For example, the ancestors of East Asians are thought to have undergone processess of selection for a number of alleles, including variants of the EDAR, ADH1B, ABCC1, and ALDH2 genes.
[32] Several phenotypical traits of characteristic of East Asians are due to a single mutation of the EDAR gene, dated to c. 35,000 years ago.
[40][41] In May 2023, scientists reported, based on genetic studies, a more complicated pathway of human evolution than previously understood.