Minimal genome
The minimal genome is a concept which can be defined as the set of genes sufficient for life to exist and propagate under nutrient-rich and stress-free conditions.
Alternatively, it may be defined as the gene set supporting life on an axenic cell culture in rich media, and it is thought what makes up the minimal genome will depend on the environmental conditions that the organism inhabits.
Therefore, if a collection of all the essential genes were put together, a minimum genome could be created artificially in a stable environment.
[2][3] In order to create a new organism a scientist must determine the minimal set of genes required for metabolism and replication.
This can be achieved by experimental and computational analysis of the biochemical pathways needed to carry out basic metabolism and reproduction.
[5] Scientifically, minimal genome projects allow the identification of the most essential genes, and the reduction of genetic complexity, making engineered strains more predictable.
Genome reduction driven by mutation and genetic drift in small and asexual populations with biases for gene deletion can be seen in symbionts and parasites, which commonly experience rapid evolution, codon reassignments, biases for AT nucleotide compositions, and elevated levels of protein misfolding which results in a heavy dependence on molecular chaperones to ensure protein functionality.
[1] These effects, which coincide with the proliferation of mobile genetic elements, pseudogenes, genome rearrangements, and chromosomal deletion are best studied and observed in more recently evolved symbionts.
The most extreme examples of genome reduction have been found in maternally transmitted endosymbionts which have experienced lengthy coevolution with their hosts and, in the process, lost a substantial amount of their cellular autonomy.
Beneficial symbionts have a greater capacity for genome reduction than do parasites, as host co-adaptation allows them to lose additional crucial genes.
[20] Organelles known as plastids in plants (including chloroplasts, chromoplasts, and leucoplasts), once free-living cyanobacteria, typically retain longer genomes on the order of 100-200kb with 80-250 genes.
[22] Examples of organisms which have experienced genome reduction include species of Buchnera, Chlamydia, Treponema, Mycoplasma, and many others.
[26] In 2006, another endosymbiont Carsonella ruddii was found with a reduced genome 160 kb in length encompassing 182 protein-coding genes.
[29] In 2011, Tremblaya princeps was found to contain an intracellular endosymbiont with a genome of 139 kb, reduced to the point that even some translation genes had been lost.
Combined, the genomes of these two symbionts can only synthesize ten amino acids, in addition to some of the machinery involved in DNA replication, transcription, and translation.
For instance, bacteriophage MS2 consists of only 3569 nucleotides (single-stranded RNA) and encodes just four proteins which overlap to make efficient use of the genome space.
[34] This concept arose as a result of a collaborative effort between National Aeronautics and Space Administration (NASA) and two scientists: Harold Morowitz and Mark Tourtellotte.
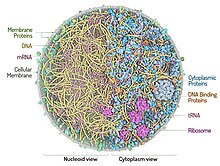
Mycoplasmas were selected as the best candidate for cell reassembly, since they are composed of a minimum set of organelles, such as a plasma membrane, ribosomes and a circular double stranded DNA.
In 1995, another laboratory from Maryland the Institute for Genomic Research (TIGR) collaborated with the teams of Johns Hopkins and the University of North Carolina.
The J. Craig Venter institute conducted these types of experiment on M. genitalium and found 382 essential genes.
The J.Craig Venter institute later started a project to create a synthetic organism named Mycoplasma laboratorium, using the minimal set genes identified from M.
Once the set of essential genetic elements are known, one can proceed to define the key pathways and core-players by modeling simulations and wet lab genome engineering.
[3] As of 1999, the two organisms upon which the ‘minimal gene set for cellular life' have been applied are: Haemophilus influenzae, and M. genitalium.
Without a fully sequenced genome it would not be possible to determine the essential minimal gene set required for survival.
[3] J. Craig Venter Institute (JCVI) conducted a study to find all the essential genes of M. genitalium through global transposon mutagenesis.
In their experiment they grew a set of Tn4001 transformed cells for many weeks and isolated the genomic DNA from these mixture of mutants.
The ultimate result of this project has now come down to constructing a synthetic organism, Mycoplasma laboratorium based on the 387 protein coding region and 43 structural RNA genes found in M.
[needs update] Researchers at the JCVI in 2010 successfully created a synthetic bacterial cell that was capable of replicating itself.
When cells are grown on minimal media, many more genes are essential as they may be needed to synthesize such nutrients (e.g. vitamins).
The numbers provided in the following table typically have been collected using rich media (but consult original references for details).