Chromosome conformation capture
Treatment of cells with 1-3% formaldehyde, for 10-30min at room temperature is most common, however, standardization for preventing high protein-DNA cross linking is necessary, as this may negatively affect the efficiency of restriction digestion in the subsequent step.
The 5C technique overcomes the junctional problems at the intramolecular ligation step and is useful for constructing complex interactions of specific loci of interest.
The pair of sequences are individually aligned to the genome, thus determining the fragments involved in that ligation event.
A number of methods use oligonucleotide capture to enrich 3C and Hi-C libraries for specific loci of interest.
[39] Single-cell adaptations of these methods, such as ChIP-seq and Hi-C can be used to investigate the interactions occurring in individual cells.
[2][46] The ChIP-loop may be useful in identifying long-range cis-interactions and trans interaction mediated through proteins since frequent DNA collisions will not occur.
[citation needed] ChIA-PET combines Hi-C with ChIP-seq to detect all interactions mediated by a protein of interest.
[47] 3C methods have led to a number of biological insights, including the discovery of new structural features of chromosomes, the cataloguing of chromatin loops, and increased understanding of transcriptional regulation mechanisms (the disruption of which can lead to disease).
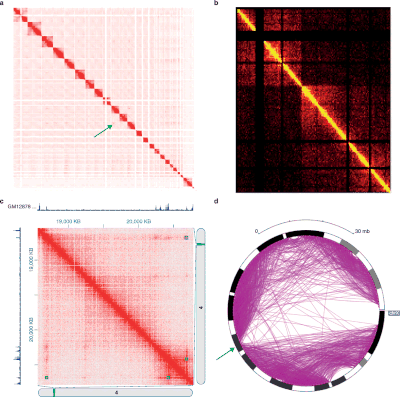
[not verified in body] These methods have revealed large-scale organization of the genome into topologically associating domains (TADs), which correlate with epigenetic markers.
The result shows that the orientation of CTCF binding motifs in an enhancer-promoter loop should be facing to each other in order for the enhancer to find its correct target.
[52] Beta thalassemia is a certain type of blood disorder caused by a deletion of LCR enhancer element.
[53][54] Holoprosencephaly is cephalic disorder caused by a mutation in the SBE2 enhancer element, which in turn weakened the production of SHH gene.
[55] PPD2 (polydactyly of a triphalangeal thumb) is caused by a mutation of ZRS enhancer, which in turn strengthened the production of SHH gene.
Fit-Hi-C [3] is a method based on a discrete binning approach with modifications of adding distance of interaction (initial spline fitting, aka spline-1) and refining the null model (spline-2).
Each eigenvector corresponds to a set of loci, which are not necessarily linearly contiguous, that share structural features.
[65] A significant confounding factor in 3C technologies is the frequent non-specific interactions between genomic loci that occur due to random polymer behavior.
Bailey et al. has identified that ZNF143 motif in the promoter regions provides sequence specificity for promoter-enhancer interactions.
[71] Moreover, they can show changes of spatial proximity for regulatory elements and their target genes, which bring deeper understanding of the structural and functional basis of the genome.