Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression.
Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs).
Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language.
In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule.
This directionality is because RNA polymerase can only add nucleotides to the 3' end of the growing mRNA chain.
[7] Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites.
In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters.
[11] The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene.
[21] However, unmethylated cytosines within 5'cytosine-guanine 3' sequences often occur in groups, called CpG islands, at active promoters.
[24] As noted in the previous section, transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a gene.
[28] The splice isoform DNMT3A2 behaves like the product of a classical immediate-early gene and, for instance, it is robustly and transiently produced after neuronal activation.
[29] Where the DNA methyltransferase isoform DNMT3A2 binds and adds methyl groups to cytosines appears to be determined by histone post translational modifications.
[30][31][32] On the other hand, neural activation causes degradation of DNMT3A1 accompanied by reduced methylation of at least one evaluated targeted promoter.
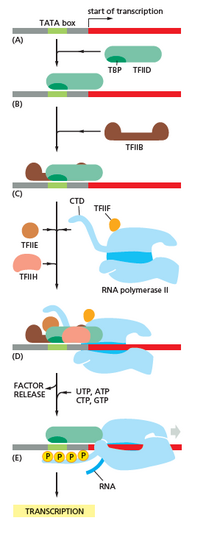
This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that thymines are replaced with uracils, and the nucleotides are composed of a ribose (5-carbon) sugar whereas DNA has deoxyribose (one fewer oxygen atom) in its sugar-phosphate backbone).
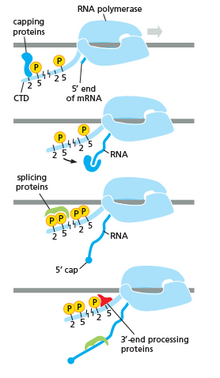
In eukaryotes, this may correspond with short pauses during transcription that allow appropriate RNA editing factors to bind.
[citation needed] Double-strand breaks in actively transcribed regions of DNA are repaired by homologous recombination during the S and G2 phases of the cell cycle.
When the hairpin forms, the mechanical stress breaks the weak rU-dA bonds, now filling the DNA–RNA hybrid.
In bacteria, the Mfd ATPase can remove a RNA polymerase stalled at a lesion by prying open its clamp.
[50] In eukayrotes, ATPase TTF2 helps to suppress the action of RNAP I and II during mitosis, preventing errors in chromosomal segregation.
[53] The regulation of transcription by processes using base excision repair and/or topoisomerases to cut and remodel the genome also increases the vulnerability of DNA to damage.
Potent, bioactive natural products like triptolide that inhibit mammalian transcription via inhibition of the XPB subunit of the general transcription factor TFIIH has been recently reported as a glucose conjugate for targeting hypoxic cancer cells with increased glucose transporter production.
Transcription factories can also be localized using fluorescence in situ hybridization or marked by antibodies directed against polymerases.
[61] A molecule that allows the genetic material to be realized as a protein was first hypothesized by François Jacob and Jacques Monod.
Severo Ochoa won a Nobel Prize in Physiology or Medicine in 1959 for developing a process for synthesizing RNA in vitro with polynucleotide phosphorylase, which was useful for cracking the genetic code.
[citation needed] Roger D. Kornberg won the 2006 Nobel Prize in Chemistry "for his studies of the molecular basis of eukaryotic transcription".
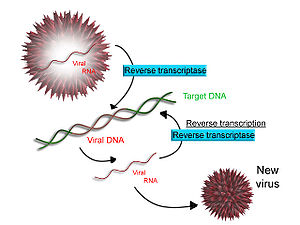
[62] Transcription can be measured and detected in a variety of ways:[citation needed] Some viruses (such as HIV, the cause of AIDS), have the ability to transcribe RNA into DNA.
The main enzyme responsible for synthesis of DNA from an RNA template is called reverse transcriptase.
[65] In the case of HIV, reverse transcriptase is responsible for synthesizing a complementary DNA strand (cDNA) to the viral RNA genome.
[citation needed] Some eukaryotic cells contain an enzyme with reverse transcription activity called telomerase.
Telomerase carries an RNA template from which it synthesizes a telomere, a repeating sequence of DNA, to the end of linear chromosomes.