Real-time polymerase chain reaction
Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence.
[1] The acronym "RT-PCR" commonly denotes reverse transcription polymerase chain reaction and not real-time PCR, but not all authors adhere to this convention.
The data thus generated can be analysed by computer software to calculate relative gene expression (or mRNA copy number) in several samples.
Older methods were used to measure mRNA abundance: differential display, RNase protection assay and northern blot.
Although this technique is still used to assess gene expression, it requires relatively large amounts of RNA and provides only qualitative or semi quantitative information of mRNA levels.
[4] Estimation errors arising from variations in the quantification method can be the result of DNA integrity, enzyme efficiency and many other factors.
[4] Real-time PCR is carried out in a thermal cycler with the capacity to illuminate each sample with a beam of light of at least one specified wavelength and detect the fluorescence emitted by the excited fluorophore.
The thermal cycler is also able to rapidly heat and chill samples, thereby taking advantage of the physicochemical properties of the nucleic acids and DNA polymerase.
This method has the advantage of only needing a pair of primers to carry out the amplification, which keeps costs down; multiple target sequences can be monitored in a tube by using different types of dyes.
The specificity of fluorescent reporter probes also prevents interference of measurements caused by primer dimers, which are undesirable potential by-products in PCR.
However, fluorescent reporter probes do not prevent the inhibitory effect of the primer dimers, which may depress accumulation of the desired products in the reaction.
[11] Unlike conventional PCR, this method avoids the previous use of electrophoresis techniques to demonstrate the results of all the samples.
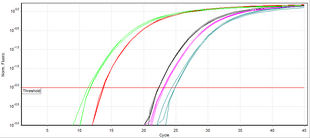
A commonly employed method of DNA quantification by real-time PCR relies on plotting fluorescence against the number of cycles on a logarithmic scale.
A threshold for detection of DNA-based fluorescence is set 3–5 times of the standard deviation of the signal noise above background.
This normalization procedure is commonly called the ΔCt-method[13] and permits comparison of expression of a gene of interest among different samples.
[14][15] Although cycle threshold analysis is integrated with many commercial software systems, there are more accurate and reliable methods of analysing amplification profile data that should be considered in cases where reproducibility is a concern.
However, theoretical analysis of polymerase chain reaction, from which MAK2 was derived, has revealed that amplification efficiency is not constant throughout PCR.
The reason for using one or more housekeeping genes is to correct non-specific variation, such as the differences in the quantity and quality of RNA used, which can affect the efficiency of reverse transcription and therefore that of the whole PCR process.
[23] However, research has shown that amplification of the majority of reference genes used in quantifying the expression of mRNA varies according to experimental conditions.
[34] The agricultural industry is constantly striving to produce plant propagules or seedlings that are free of pathogens in order to prevent economic losses and safeguard health.
Discrimination between the DNA of the pathogen and the plant is based on the amplification of ITS sequences, spacers located in ribosomal RNA gene's coding area, which are characteristic for each taxon.
Specific primers are used that amplify not the transgene but the promoter, terminator or even intermediate sequences used during the process of engineering the vector.
[37][38] Viruses can be present in humans due to direct infection or co-infections which makes diagnosis difficult using classical techniques and can result in an incorrect prognosis and treatment.
[41] Real-time PCR has also brought the quantization of human cytomegalovirus (CMV) which is seen in patients who are immunosuppressed following solid organ or bone marrow transplantation.