Restriction site associated DNA markers
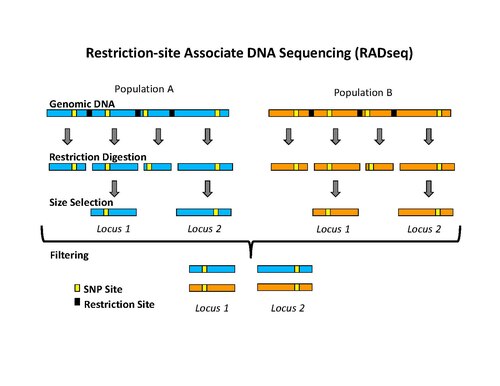
[2] The initial procedure to isolate RAD tags involved digesting DNA with a particular restriction enzyme, ligating biotinylated adapters to the overhangs, randomly shearing the DNA into fragments much smaller than the average distance between restriction sites, and isolating the biotinylated fragments using streptavidin beads.
[1][6][7] More recently, the RAD tag isolation procedure has been modified for use with high-throughput sequencing on the Illumina platform, which has the benefit of greatly reduced raw error rates and high throughput.
[1][6][7] Due to the low sensitivity of microarrays, this approach can only detect either DNA sequence polymorphisms that disrupt restriction sites and lead to the absence of RAD tags or substantial DNA sequence polymorphisms that disrupt RAD tag hybridization.
They confirmed the utility of RAD markers by identifying recombination breakpoints in D. melanogaster and by detecting QTLs in threespine sticklebacks.
[10][11] By adding a second restriction enzyme, replacing the random shearing, and a tight DNA size selection step it is possible to perform low-cost population genotyping.
This simple and cost-effective approach allows sequencing of orthologous loci even from highly degraded DNA samples, opening new avenues of research in the field of museomics.
Another advantage of the method is not relying on the restriction site presence, improving among-sample loci coverage.