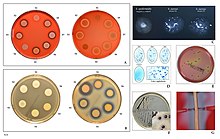
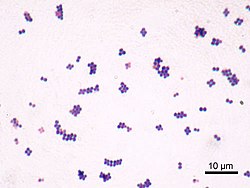
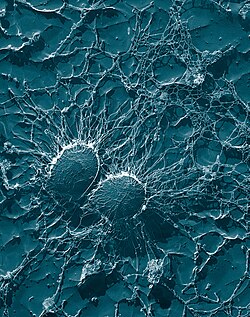
Staphylococcus aureus
[9] In 1880, Alexander Ogston, a Scottish surgeon, discovered that Staphylococcus can cause wound infections after noticing groups of bacteria in pus from a surgical abscess during a procedure he was performing.
Mobile genetic elements that are common in S. aureus include bacteriophages, pathogenicity islands, plasmids, transposons, and staphylococcal cassette chromosomes.
They also determined some genetic variations in humans that lead to an increased ability for S. aureus to colonize, notably a polymorphism in the glucocorticoid receptor gene that results in larger corticosteroid production.
[23] Natural genetic transformation is a reproductive process involving DNA transfer from one bacterium to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination.
Joint replacements put a person at particular risk of septic arthritis, staphylococcal endocarditis (infection of the heart valves), and pneumonia.
[33][32] S. aureus is believed to exploit defects in the skin barrier of persons with atopic dermatitis, triggering cytokine expression and therefore exacerbating symptoms.
[44] Infection is generally associated with breaks in the skin or mucosal membranes due to surgery, injury, or use of intravascular devices such as cannulas, hemodialysis machines, or hypodermic needles.
[69] Toxins that play a role in intraspecies competition confers an advantage by promoting successful colonisation in polymicrobial communities such as the nasopharynx and lung by outcompeting lesser strains.
[69] There are also T7 effector proteins that play role a in pathogenesis, for example mutational studies of S. aureus have suggested that EsxB and EsxC contribute to persistent infection in a murine abscess model.
[71] Altogether, T7SS and known secreted effector proteins are a strategy of pathogenesis by improving fitness against competitor S. aureus species as well as increased virulence via evading the innate immune system and optimising persistent infections.
[84] The S. aureus biofilm is embedded in a glycocalyx slime layer and can consist of teichoic acids, host proteins, extracellular DNA (eDNA) and sometimes polysaccharide intercellular antigen (PIA).
Though the exact mechanism of resistance is unknown, S. aureus biofilms have increased growth under the presence of cytokines produced by the host immune response.
Two of these genes include rocD and gudB, which encode for the enzyme's ornithine-oxo-acid transaminase and glutamate dehydrogenase, which are important for amino acid metabolism.
Transpeptidases, such as the sortases responsible for anchoring factors like protein A to the staphylococcal peptidoglycan, are being studied in hopes of developing new antibiotics to target MRSA infections.
This pigment acts as a virulence factor, primarily by being a bacterial antioxidant which helps the microbe evade the reactive oxygen species which the host immune system uses to kill pathogens.
[92] In fact, because of similarities in the pathways for biosynthesis of staphyloxanthin and human cholesterol, a drug developed in the context of cholesterol-lowering therapy was shown to block S. aureus pigmentation and disease progression in a mouse infection model.
Recent genetic advances have enabled reliable and rapid techniques for the identification and characterization of clinical isolates of S. aureus in real time.
The S. aureus fragments then transition down the gel, producing specific band patterns that are later compared with other isolates in hopes of identifying related strains.
[101] Although this technique is often inexpensive and less time-consuming, the chance of losing discriminatory power making it hard to differentiate between MLST clonal complexes exemplifies a crucial limitation.
An antibiotic derived from some Penicillium fungal species, penicillin inhibits the formation of peptidoglycan cross-linkages that provide the rigidity and strength in a bacterial cell wall.
Adjunctive rifampicin has been historically used in the management of S aureus bacteraemia, but randomised controlled trial evidence has shown this to be of no overall benefit over standard antibiotic therapy.
Indeed, the original Petri dish on which Alexander Fleming of Imperial College London observed the antibacterial activity of the Penicillium fungus was growing a culture of S. aureus.
[107] One topical agent that is prescribed is mupirocin, a protein synthesis inhibitor that is produced naturally by Pseudomonas fluorescens and has seen success for treatment of S. aureus nasal carriage.
Penicillinase-resistant β-lactam antibiotics, such as methicillin, nafcillin, oxacillin, cloxacillin, dicloxacillin, and flucloxacillin are able to resist degradation by staphylococcal penicillinase.
[99] Resistance is conferred by the mecA gene, which codes for an altered penicillin-binding protein (PBP2a or PBP2') that has a lower affinity for binding β-lactams (penicillins, cephalosporins, and carbapenems).
[109] One study suggests that MRSA sacrifices virulence, for example, toxin production and invasiveness, for survival and creation of biofilms[110] Aminoglycoside antibiotics, such as kanamycin, gentamicin, streptomycin, were once effective against staphylococcal infections until strains evolved mechanisms to inhibit the aminoglycosides' action, which occurs via protonated amine and/or hydroxyl interactions with the ribosomal RNA of the bacterial 30S ribosomal subunit.
[citation needed] Glycopeptide resistance is typically mediated by acquisition of the vanA gene, which originates from the Tn1546 transposon found in a plasmid in enterococci and codes for an enzyme that produces an alternative peptidoglycan to which vancomycin will not bind.
[125] Among the various mechanisms that MRSA acquires to elude antibiotic resistance (e.g., drug inactivation, target alteration, reduction of permeability) there is also the overexpression of efflux pumps.
[citation needed] By directly modulating efflux pumps' activity or decreasing their expression, it may be possible to modify the resistant phenotype and restore the effectiveness of existing antibiotics.
[153] The vaccine underwent clinical trial until June 2019, with results published in September 2020, that did not demonstrate a significant reduction in Postoperative Bloodstream Infection after Surgery.