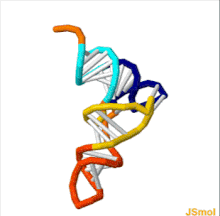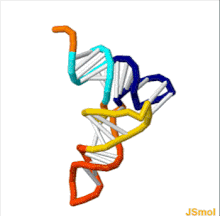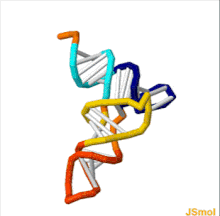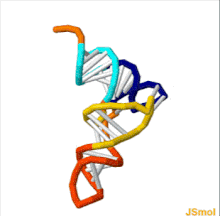
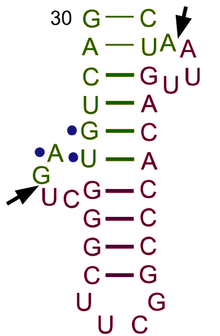
Transfer RNA
This is first transformed into mRNA, then tRNA specifies which three-nucleotide codon from the genetic code corresponds to which amino acid.
[3] Each mRNA codon is recognized by a particular type of tRNA, which docks to it along a three-nucleotide anticodon, and together they form three complementary base pairs.
These unusual bases sometimes affect the tRNA's interaction with ribosomes and sometimes occur in the anticodon to alter base-pairing properties.
Each tRNA has a distinct anticodon triplet sequence that can form 3 complementary base pairs to one or more codons for an amino acid.
Frequently, the first nucleotide of the anticodon is one not found on mRNA: inosine, which can hydrogen bond to more than one base in the corresponding codon position.
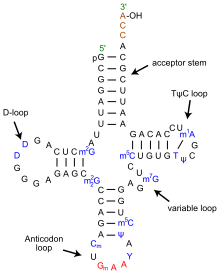
Recognition of the appropriate tRNA by the synthetases is not mediated solely by the anticodon, and the acceptor stem often plays a prominent role.
An amidotransferase then converts the acid side chain of the glutamate to the amide, forming the correctly charged gln-tRNA-Gln.
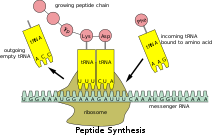
In addition, the ribosome has two other sites for tRNA binding that are used during mRNA decoding or during the initiation of protein synthesis.
[25] The binding proteins like L27, L2, L14, L15, L16 at the A- and P- sites have been determined by affinity labeling by A. P. Czernilofsky et al. (Proc.
Once translation initiation is complete, the first aminoacyl tRNA is located in the P/P site, ready for the elongation cycle described below.
Once mRNA decoding is complete, the aminoacyl-tRNA is bound in the A/A site and is ready for the next peptide bond[27] to be formed to its attached amino acid.
For example, the nematode worm C. elegans, a commonly used model organism in genetics studies, has 29,647 genes in its nuclear genome,[28] of which 620 code for tRNA.
[36][37] The phenomenon of multiple nuclear copies of mitochondrial tRNA (tRNA-lookalikes) has been observed in many higher organisms from human to the opossum[38] suggesting the possibility that the lookalikes are functional.
[33] The HGNC, in collaboration with the Genomic tRNA Database (GtRNAdb) and experts in the field, has approved unique names for human genes that encode tRNAs.
[41] The reasons why tRNA genes have been lost during evolution remains under debate but may relate improving resistance to viral infection.
The top half may have evolved first including the 3′-terminal genomic tag which originally may have marked tRNA-like molecules for replication in early RNA world.
To generate type II tRNAs, a single internal 9 nucleotide deletion occurred within ligated acceptor stems (CCGCCGCGCGGCGG goes to GGCGG).
To generate type I tRNAs, an additional, related 9 nucleotide deletion occurred within ligated acceptor stems within the variable loop region (CCGCCGCGCGGCGG goes to CCGCC).
tRNA-derived fragments (or tRFs) are short molecules that emerge after cleavage of the mature tRNAs or the precursor transcript.
[55] There are at least four structural types of tRFs believed to originate from mature tRNAs, including the relatively long tRNA halves and short 5'-tRFs, 3'-tRFs and i-tRFs.
[57] tRFs appear to play a role in RNA interference, specifically in the suppression of retroviruses and retrotransposons that use tRNA as a primer for replication.
[58] tRFs have multiple dependencies and roles; such as exhibiting significant changes between sexes, among races and disease status.
[55][59][60] Functionally, they can be loaded on Ago and act through RNAi pathways,[53][56][61] participate in the formation of stress granules,[62] displace mRNAs from RNA-binding proteins[63] or inhibit translation.
Functionally, tRFs are associated with viral infection,[65] cancer,[56] cell proliferation [57] and also with epigenetic transgenerational regulation of metabolism.
In 1990, tRNAfMet2CUA (modified from the tRNAfMet2CAU gene metY) was inserted into E. coli, causing it to initiate protein synthesis at the UAG stop codon, as long as it is preceded by a strong Shine-Dalgarno sequence.
Some pre-tRNAs contain introns that are spliced, or cut, to form the functional tRNA molecule;[80] in bacteria these self-splice, whereas in eukaryotes and archaea they are removed by tRNA-splicing endonucleases.
[81] Eukaryotic pre-tRNA contains bulge-helix-bulge (BHB) structure motif that is important for recognition and precise splicing of tRNA intron by endonucleases.
[84] A notable exception is in the archaeon Nanoarchaeum equitans, which does not possess an RNase P enzyme and has a promoter placed such that transcription starts at the 5′ end of the mature tRNA.
[100] Similarly, hepatitis E virus requires a tRNA landscape that substantially differs from that associated with uninfected cells.
[101] Hence, inhibition of aminoacylation of specific tRNA species is considered a promising novel avenue for the rational treatment of a plethora of diseases.