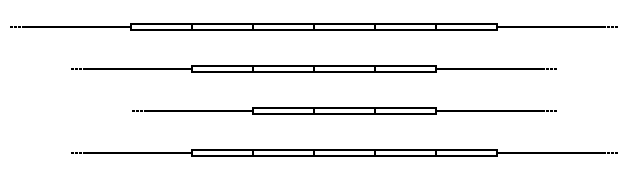
Variable number tandem repeat
Flanking regions are segments of repetitive sequence (shown here as thin lines), allowing the VNTR blocks to be extracted with restriction enzymes and analyzed by RFLP, or amplified by the polymerase chain reaction (PCR) technique and their size determined by gel electrophoresis.
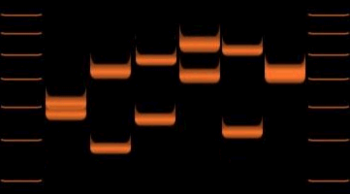
When removed from surrounding DNA by the PCR or RFLP methods, and their size determined by gel electrophoresis or Southern blotting, they produce a pattern of bands unique to each individual.
When tested with a group of independent VNTR markers, the likelihood of two unrelated individuals' having the same allelic pattern is extremely low.
In analyzing VNTR data, two basic genetic principles can be used: Repetitive DNA, representing over 40% of the human genome, is arranged in a bewildering array of patterns.
VNTRs are a type of minisatellite in which the size of the repeat sequence is generally ten to one hundred base pairs.