Virtual karyotype
Conventional karyotypes can assess the entire genome for changes in chromosome structure and number, but the resolution is relatively coarse, with a detection limit of 5-10Mb.
[citation needed] Virtual karyotypes can be performed on germline samples for constitutional disorders,[5][6] and clinical testing is available from dozens of CLIA certified laboratories (genetests.org).
Despite the diversity of platforms, ultimately they all use genomic DNA from disrupted cells to recreate a high resolution karyotype in silico.
[citation needed] Autozygous segments and uniparental disomy (UPD) are diploid/'copy neutral' genetic findings and therefore are only detectable by SNP-based arrays.
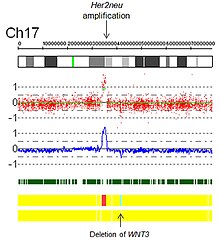
[citation needed] Figure 7 is a SNP array virtual karyotype from a colorectal carcinoma demonstrating deletions, gains, amplifications, and acquired UPD (copy neutral LOH).
Clinical utility varies and appropriateness is best determined by an oncologist or pathologist in consultation with the laboratory director of the lab performing the virtual karyotype.
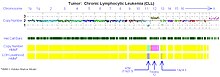
[12][34][35][36][37] In addition, many studies using array-based karyotyping have identified 'atypical deletions' missed by the standard FISH probes and acquired uniparental disomy at key loci for prognostic risk in CLL.
[40] Avet-Loiseau, et al. in Journal of Clinical Oncology, used SNP array karyotyping of 192 multiple myeloma (MM) samples to identify genetic lesions associated with prognosis, which were then validated in a separate cohort (n = 273).
Therefore, FISH for this translocation should also be performed if using SNP arrays to detect genome-wide copy number alterations of prognostic significance in MM.
[citation needed] Array-based karyotyping of 260 medulloblastomas by Pfister S, et al. resulted in the following clinical subgroups based on cytogenetic profiles:[42] The 1p/19q co-deletion is considered a "genetic signature" of oligodendroglioma.
Cytogenetics, the study of characteristic large changes in the chromosomes of cancer cells, has been increasingly recognized as an important predictor of outcome in acute lymphoblastic leukemia (ALL).
Cytogenetics play a decisive role in the World Health Organization's classification-based International Prognostic Scoring System (IPSS) for MDS.
[57][58] In a comparison of metaphase cytogenetics, FISH panel, and SNP array karyotyping for MDS, it was found that each technique provided a similar diagnostic yield.
[59] Acquired UPD, which is not detectable by FISH or cytogenetics, has been reported at several key loci in MDS using SNP array karyotyping, including deletion of 7/7q.
[60][61] Philadelphia chromosome–negative myeloproliferative neoplasms (MPNs) including polycythemia vera, essential thrombocythemia, and primary myelofibrosis show an inherent tendency for transformation into leukemia (MPN-blast phase), which is accompanied by acquisition of additional genomic lesions.
In a study of 159 cases,[62] SNP-array analysis was able to capture practically all cytogenetic abnormalities and to uncover additional lesions with potentially important clinical implications.
[citation needed] Identification of biomarkers in colorectal cancer is particularly important for patients with stage II disease, where less than 20% have tumor recurrence.
Colorectal cancers are classified into specific tumor phenotypes based on molecular profiles[63] which can be integrated with the results of other ancillary tests, such as microsatellite instability testing, IHC, and KRAS mutation status: Malignant rhabdoid tumors are rare, highly aggressive neoplasms found most commonly in infants and young children.
[citation needed] SNP array karyotyping can be used to distinguish, for example, a medulloblastoma with an isochromosome 17q from a primary rhabdoid tumor with loss of 22q11.2.
[64] The most important genetic alteration associated with poor prognosis in uveal melanoma is loss of an entire copy of Chromosome 3 (Monosomy 3), which is strongly correlated with metastatic spread.
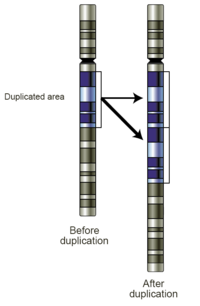
[66] In rare instances, monosomy 3 tumors may duplicate the remaining copy of the chromosome to return to a disomic state referred to as isodisomy.
For karyotypes obtained from SNP-based arrays, tetraploidy can often be inferred from the maintenance of heterozygosity within a region of apparent copy number loss.
[22] Low-level mosaicism or small subclones may not be detected by virtual karyotypes because the presence of normal cells in the sample will dampen the signal from the abnormal clone.