Omics
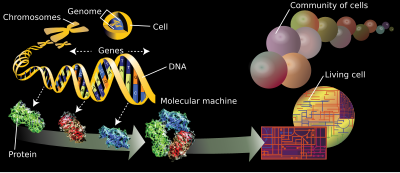
[1][2] The branches of science known informally as omics are various disciplines in biology whose names end in the suffix -omics, such as genomics, proteomics, metabolomics, metagenomics, phenomics and transcriptomics.
The related suffix -ome is used to address the objects of study of such fields, such as the genome, proteome or metabolome respectively.
"Omes" can provide an easy shorthand to encapsulate a field; for example, an interactomics study is clearly recognisable as relating to large-scale analyses of gene-gene, protein-protein, or protein-ligand interactions.
Researchers are rapidly taking up omes and omics, as shown by the explosion of the use of these terms in PubMed since the mid-1990s.
The microbiome is a microbial community occupying a well-defined habitat with distinct physio-chemical properties.
Foodomics was defined by Alejandro Cifuentes in 2009 as "a discipline that studies the food and nutrition domains through the application and integration of advanced omics technologies to improve consumer’s well-being, health, and knowledge.
Inspired by foundational questions in evolutionary biology, a Harvard team around Jean-Baptiste Michel and Erez Lieberman Aiden created the American neologism culturomics for the application of big data collection and analysis to cultural studies.