1000 Genomes Project
The 1000 Genomes Project (1KGP), taken place from January 2008 to 2015, was an international research effort to establish the most detailed catalogue of human genetic variation at the time.
Scientists planned to sequence the genomes of at least one thousand anonymous healthy participants from a number of different ethnic groups within the following three years, using advancements in newly developed technologies.
Patterns of DNA polymorphisms can be used to reliably detect signatures of selection and may help to identify genes that might underlie variation in disease resistance or drug metabolism.
[12][13] Such insights could improve understanding of phenotypic variations, genetic disorders and Mendelian inheritance and their effects on survival and/or reproduction of different human populations.
[15] Secondary goals included the support of better SNP and probe selection for genotyping platforms in future studies and the improvement of the human reference sequence.
[15] The human genome consists of approximately 3 billion DNA base pairs and is estimated to carry around 20,000 protein coding genes.
In designing the study the consortium needed to address several critical issues regarding the project metrics such as technology challenges, data quality standards and sequence coverage.
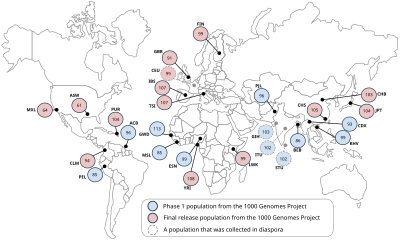
The following populations will be included in the study: Yoruba in Ibadan (YRI), Nigeria; Japanese in Tokyo (JPT); Chinese in Beijing (CHB); Utah residents with ancestry from northern and western Europe (CEU); Luhya in Webuye, Kenya (LWK); Maasai in Kinyawa, Kenya (MKK); Toscani in Italy (TSI); Peruvians in Lima, Peru (PEL); Gujarati Indians in Houston (GIH); Chinese in metropolitan Denver (CHD); people of Mexican ancestry in Los Angeles (MXL); and people of African ancestry in the southwestern United States (ASW).