Algae DNA barcoding
It is conducted using DNA or RNA followed by amplification and sequencing of specific, conserved regions in the diatom genome followed by taxonomic assignment.
[3] Using the V4 hypervariable region of the ribosomal small subunit DNA (SSU rDNA), DNA-based identification was found to be more efficient than the classical morphology-based approach.
Applying the DNA barcoding concept to diatoms promises great potential to resolve the problem of inaccurate species identification and thus facilitate analyses of the biodiversity of environmental samples.
[14] Molecular methods based on the NGS technology almost always leads to a higher number of identified taxa whose presence could subsequently be verified by light microscopy.
[4] Results of this study provides evidence that eDNA barcoding of diatoms is suitable for water quality assessment and could complement or improve traditional methods.
Moreover, DNA-based identification allows extending the range of potential bioindicators, including the inconspicuous taxonomic groups that could be highly sensitive or tolerant to particular stressors.
It has also been shown that taxonomic identification with DNA barcoding is not accurate above species level, to discriminate varieties for example (reference missing).
For example, Achnanthidium minutissimum has a small biovolume, and thus will generate less copies of the rbcL fragment (located in the chloroplast) than larger species.
Several DNA markers, belonging to the nuclear, mitochondrial, and chloroplast genomes (rbcL, COI, ITS+5.8S, SSU, 18S...), have been designed and successfully used for diatoms identification with NGS.
The rbcl gene is used for taxonomy studies (Trobajo et al. 2009) which benefits include that rarely any intragenomic variation and they are very easily aligned and compared.
[29] Diatoms are used as an indicator of ecosystem health in freshwaters because they are ubiquitous, directly affected by the changes in physico-chemical parameters and show a better relationship with environmental variables than other taxa e.g. invertebrates, giving a better overall picture of water quality.
[30] Over the recent years, researchers have developed and standardised the tools for the metabarcoding and sequencing of diatoms, to complement the traditional assessment using microscopy, opening up a new avenue of biomonitoring for aquatic systems.
[5] Many studies have shown that metabarcoding and HTS (high-throughput sequencing) can be utilized to estimate the quality status and diversity in freshwaters.
As part of the Environment Agency, Kelly et al.[32] has developed a DNA-based metabarcoding approach to assess diatom communities in rivers for the UK.
Vasselon et al.[34] also applied DNA metabarcoding of diatoms communities to the monitoring network of rivers on the tropical Island Mayotte (French DOM-TOM).
DNA barcoding and metabarcoding can be used to establish molecular metrics and indices, which potentially provide conclusions broadly similar to those of the traditional approaches about the ecological and environmental status of aquatic ecosystems.
Over the past 2 decades certain standards for DNA barcoding with the aim of species identification have been developed for each of the main groups of macroalgae.
The barcodes typically differ between the 3 main groups of macroalgae (red, green and brown) because their evolutionary heritage is very diverse.
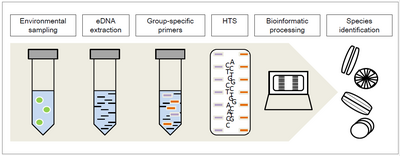

of aquatic ecosystems
