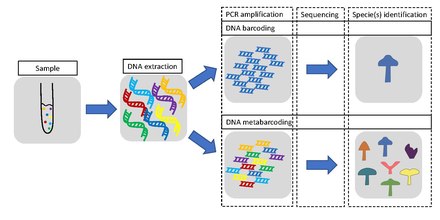
DNA barcoding
The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called "sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database.
The most commonly used barcode region for animals and some protists is a portion of the cytochrome c oxidase I (COI or COX1) gene, found in mitochondrial DNA.
Calling the profiles "barcodes", Hebert et al. envisaged the development of a COI database that could serve as the basis for a "global bioidentification system".
The eDNA method is applied on most sample types, like water, sediment, soil, animal feces, stomach content or blood from e.g.
A good DNA barcode should have low intra-specific and high inter-specific variability[12] and possess conserved flanking sites for developing universal PCR primers for wide taxonomic application.
Multi-locus markers such as ribosomal internal transcribed spacers (ITS DNA) along with matK, rbcL, trnH or other genes have also been used for species identification.
[25] Some studies suggest COI,[26] type II chaperonin (cpn60)[27] or β subunit of RNA polymerase (rpoB)[28] also could serve as bacterial DNA barcodes.
In the case of macro- and many microorganisms (such as algae), these reference libraries require detailed documentation (sampling location and date, person who collected it, image, etc.)
In some cases, due to the incompleteness of reference databases, identification can only be achieved at higher taxonomic levels, such as assignment to a family or class.
[53] DNA barcode markers can be applied to address basic questions in systematics, ecology, evolutionary biology and conservation, including community assembly, species interaction networks, taxonomic discovery, and assessing priority areas for environmental protection.
DNA barcoding has great applicability in identification of larvae for which there are generally few diagnostic characters available, and in association of different life stages (e.g. larval and adult) in many animals.
[63] Smith et al. (2007) used cytochrome c oxidase I DNA barcodes for species identification of the 20 morphospecies of Belvosia parasitoid flies (Diptera: Tachinidae) reared from caterpillars (Lepidoptera) in Area de Conservación Guanacaste (ACG), northwestern Costa Rica.
[65] DNA barcoding and metabarcoding can be useful in diet analysis studies,[66] and is typically used if prey specimens cannot be identified based on morphological characters.
[51][72] In fecal samples or highly digested stomach contents, it is often not possible to distinguish tissue from single species, and therefore metabarcoding can be applied instead.
Unknown animal or plant samples at crime scenes can be found, collected, and identified, in hopes of linking it to a suspect and getting a conviction.
DNA barcoding allows the resolution of taxa from higher (e.g. family) to lower (e.g. species) taxonomic levels, that are otherwise too difficult to identify using traditional morphological methods, like e.g. identification via microscopy.
This study found different response patterns of 12 molecular distinct OTUs to stressors which may change the consensus that this mayfly is sensitive to pollution.
Another factor might be the behavior of the target species, e.g. fish can have seasonal changes of movements, crayfish or mussels will release DNA in larger amounts just at certain times of their life (moulting, spawning).
[85] Importantly, DNA barcodes can also be used to create interim taxonomy, in which case OTUs can be used as substitutes for traditional Latin binomials – thus significantly reducing dependency on fully populated reference databases.
Several studies have highlighted the possibility to use mitochondria-enriched samples [93][94] or PCR-free approaches to avoid these biases, but as of 2018[update], the DNA metabarcoding technique is still based on the sequencing of amplicons.
[96] Another criticism of DNA barcoding is its limited efficiency for accurate discrimination below species level (for example, to distinguish between varieties), for hybrid detection, and that it can be affected by evolutionary rates[citation needed].
The most important cause is probably the incompleteness and lack of accuracy of the molecular reference databases preventing a correct taxonomic assignment of eDNA sequences.
Taxa not present in reference databases will not be found by eDNA, and sequences linked to a wrong name will lead to incorrect identification.
[102][103][104][105][106]This is enabled by the use of third-generation sequencing platforms including PacBio (Sequel I/II) by Pacific Biosciences and MinION, PromethION by Oxford Nanopore Technology.
As compared to Sanger sequencing, megabarcoding is faster and cheaper, allowing for the large-scale generation of DNA barcodes for thousands of species.
The main difference between the approaches is that metabarcoding, in contrast to barcoding, does not focus on one specific organism, but instead aims to determine species composition within a sample.
The metabarcoding procedure, like general barcoding, covers the steps of DNA extraction, PCR amplification, sequencing and data analysis.
Metabarcoding has the potential to complement biodiversity measures, and even replace them in some instances, especially as the technology advances and procedures gradually become cheaper, more optimized and widespread.
[118][119] DNA metabarcoding applications include Biodiversity monitoring in terrestrial and aquatic environments, Paleontology and ancient ecosystems, Plant-pollinator interactions, Diet analysis and Food safety.
[120] However, there are current joined attempts, like e.g. the EU COST network DNAqua-Net, to move forward by exchanging experience and knowledge to establish best-practice standards for biomonitoring.