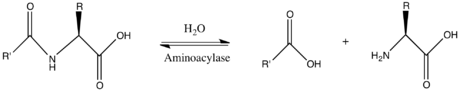
Aminoacylase
[3][5] The negatively charged hydroxide ion is nucleophilic and attacks the electrophilic carbonyl carbon of the substrate's acyl group.
[5] The exact mechanism after this point is unknown, with one possibility being that the carbonyl then reforms, breaks the amide bond, and forms the two products.
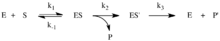
[6] This reaction sequence is an example of Michaelis–Menten kinetics, allowing one to determine KM, Kcat, Vmax, turnover number, and substrate specificity through classic Michaelis-Menten enzyme experiments.
N-acetyl-L-glutamate is an allosteric activator of carbamoyl phosphate synthetase, a crucial enzyme that commits NH4+ molecules to the urea cycle.
[7] The urea cycle gets rid of excess ammonia (NH4+) in the body, a process that must be up-regulated during times of increased protein catabolism, as amino acid breakdown produces large amounts of NH4+.
[7] Aminoacylase is up-regulated during times of nutrient deficit or starvation, causing N-acetyl-L-glutamate breakdown, which down-regulates carbamoyl phosphate synthetase and the rest of the urea cycle.
This response is evolutionarily advantageous, since a nutrient deficit means there isn't as much NH4+ that needs to be disposed of and since the body wants to salvage as many amino acids as it can.
[4] Porcine aminoacylase 1 is composed of two identical heterodimeric subunits each consisting of 406 amino acids, with acetylalanine at the N-terminus of each.
[1][4][22] It can be inferred from this data that these two enzymes evolved from a common ancestral protein, retaining function but diverging in structure over time.